Views
Single View

We characterize a view as a set of one or multiple aligned tracks, with at most one two-axis track. In a single view, we cannot visualize multiple scales and foci.
3D Genome Browser 3dViewer
3dViewer AliView
AliView Alvis
Alvis BactoGenie
BactoGenie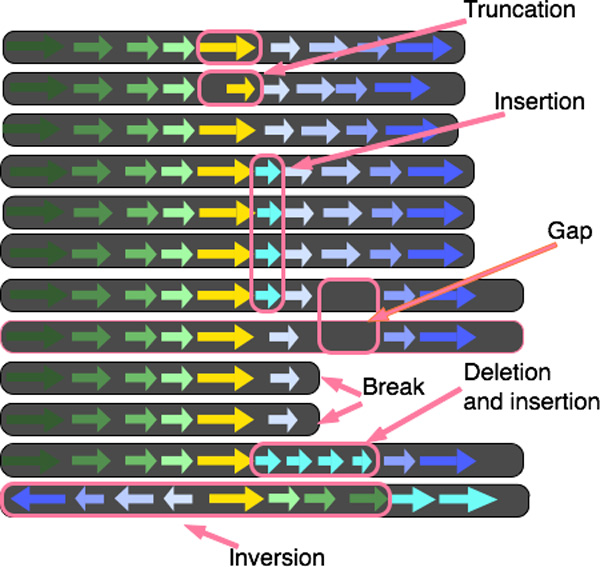 CGView
CGView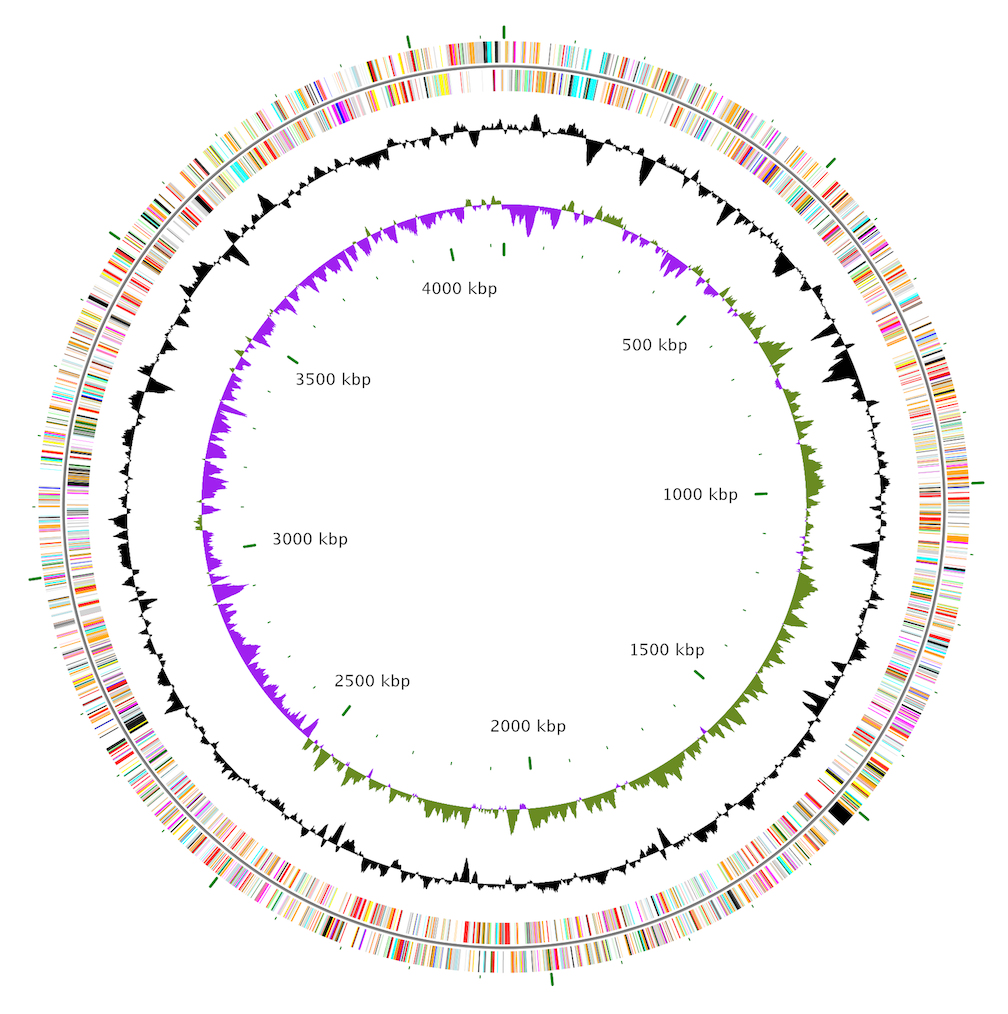 CGView Server
CGView Server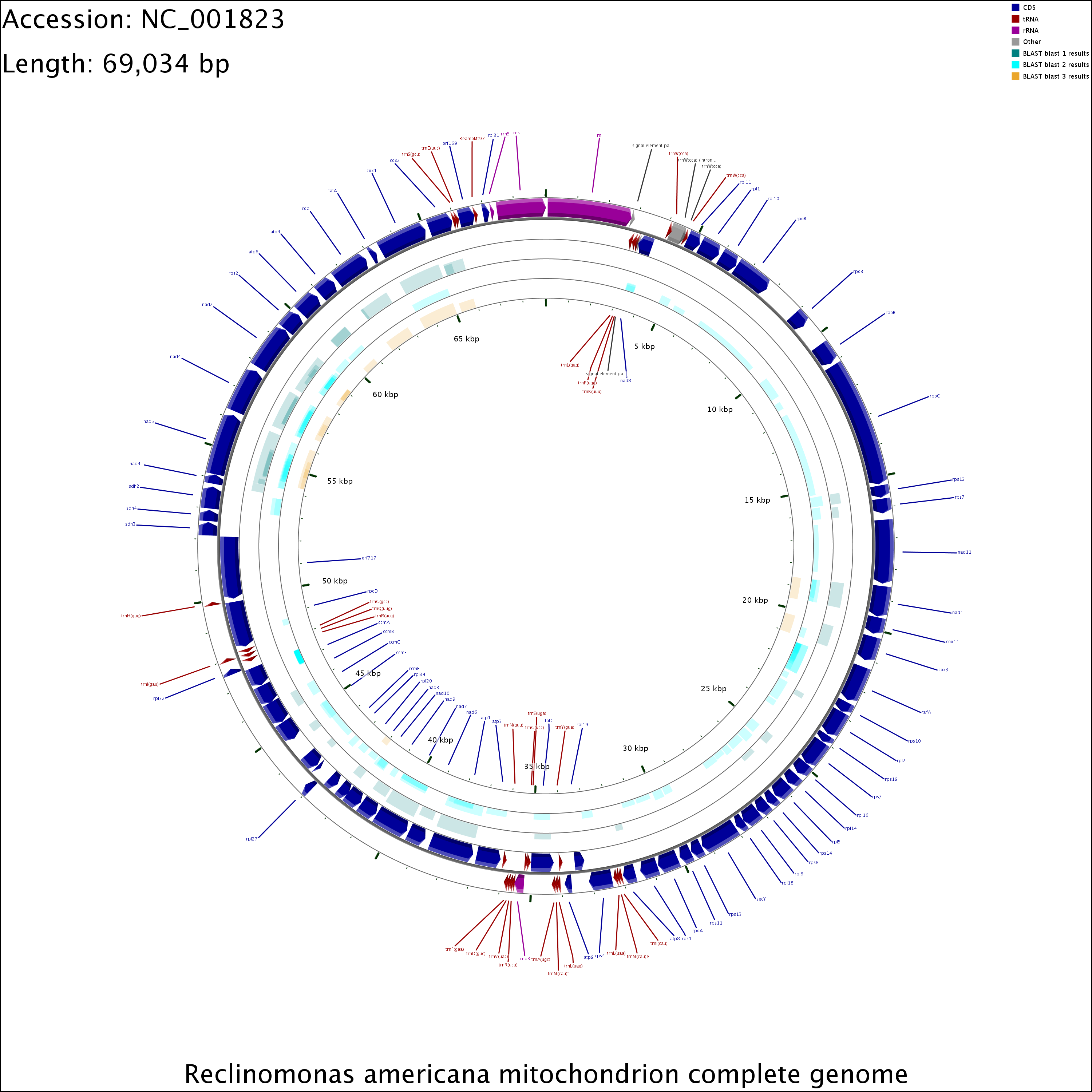 Cinteny
Cinteny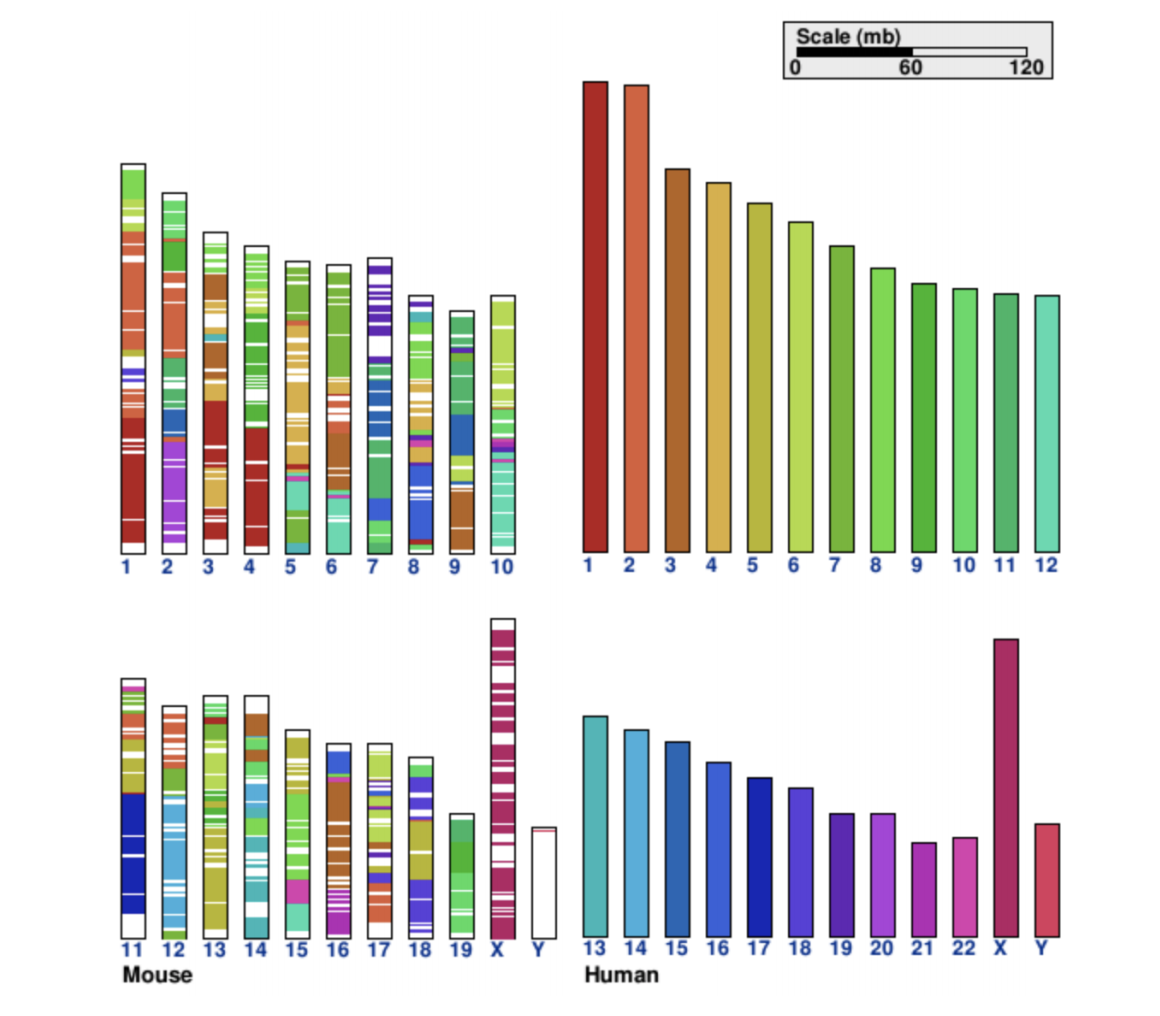 Circos
Circos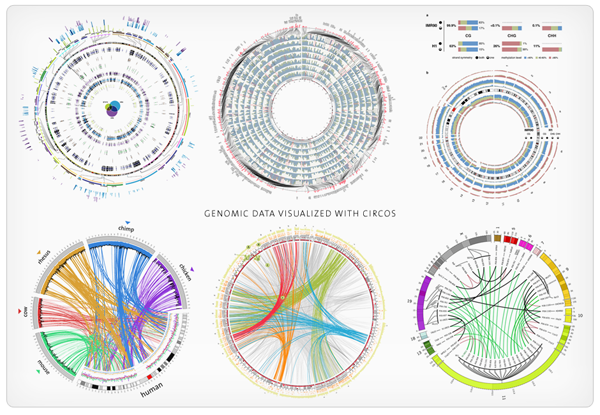 Civi
Civi ClicO Free Service
ClicO Free Service CNVkit
CNVkit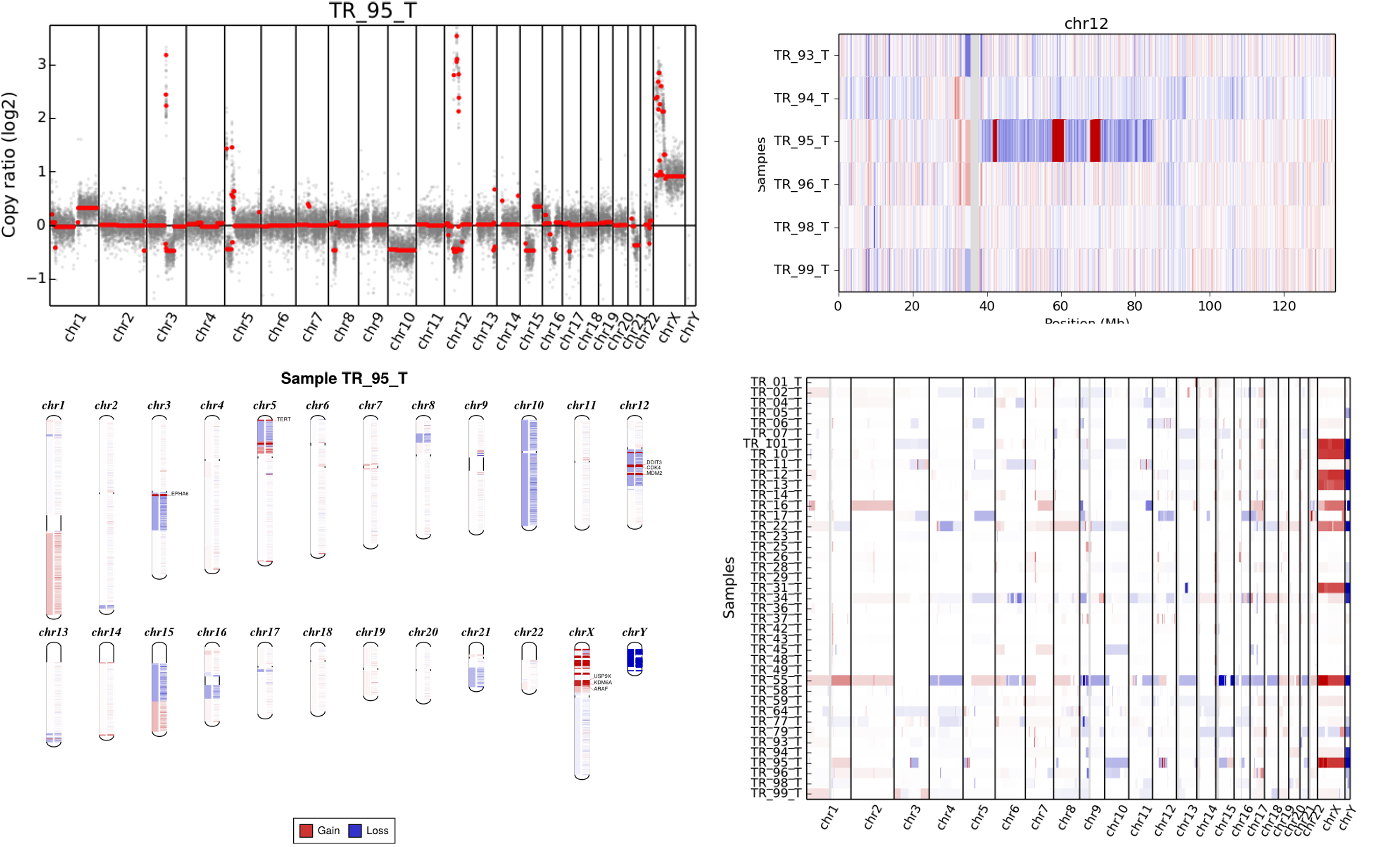 Combo
Combo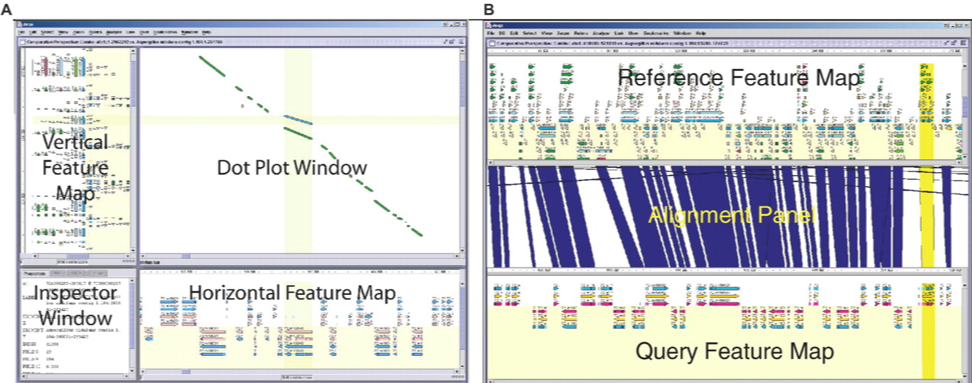 CRISPResso2
CRISPResso2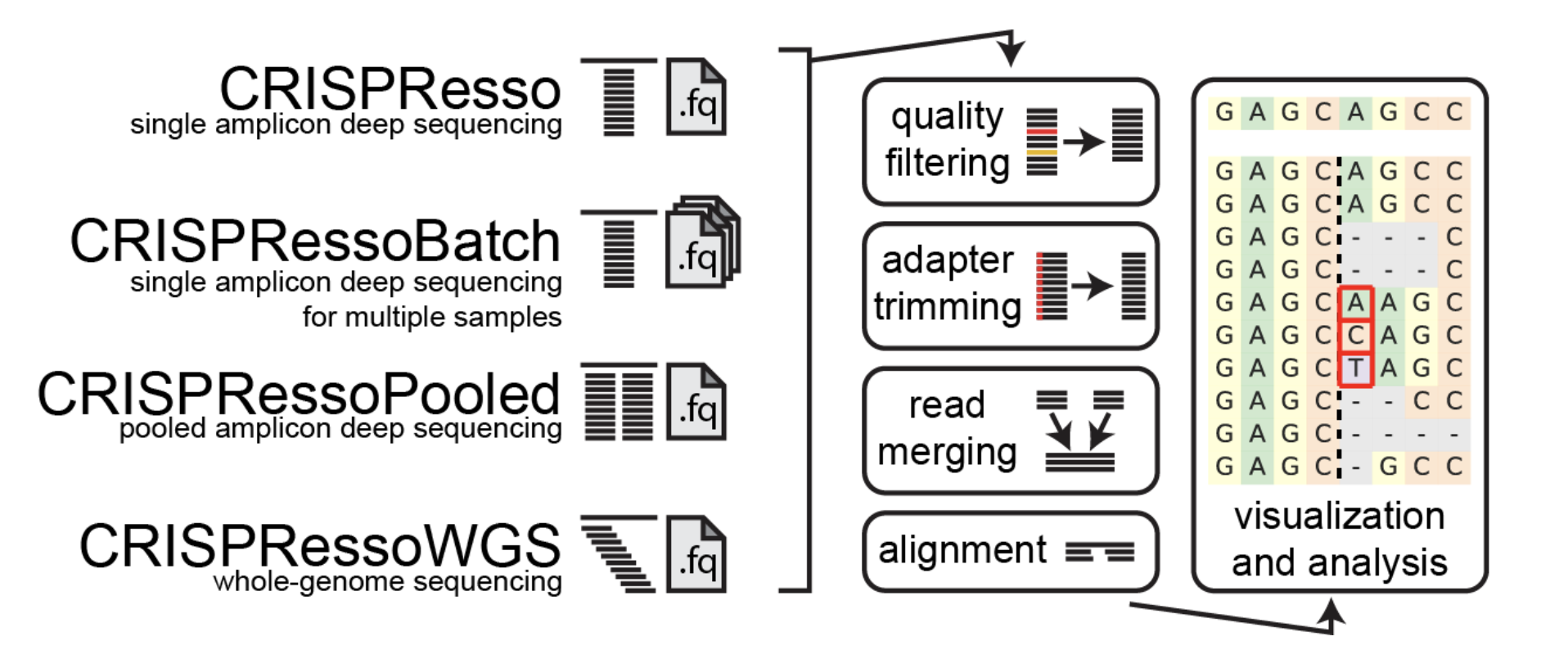 Dalliance
Dalliance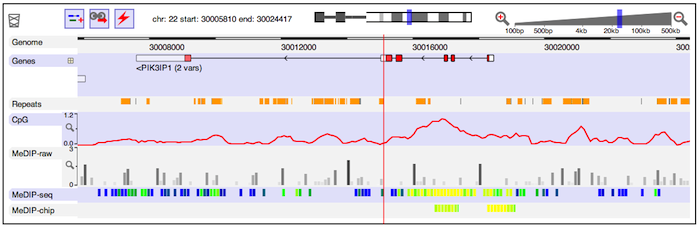 Deep Motif Dashboard
Deep Motif Dashboard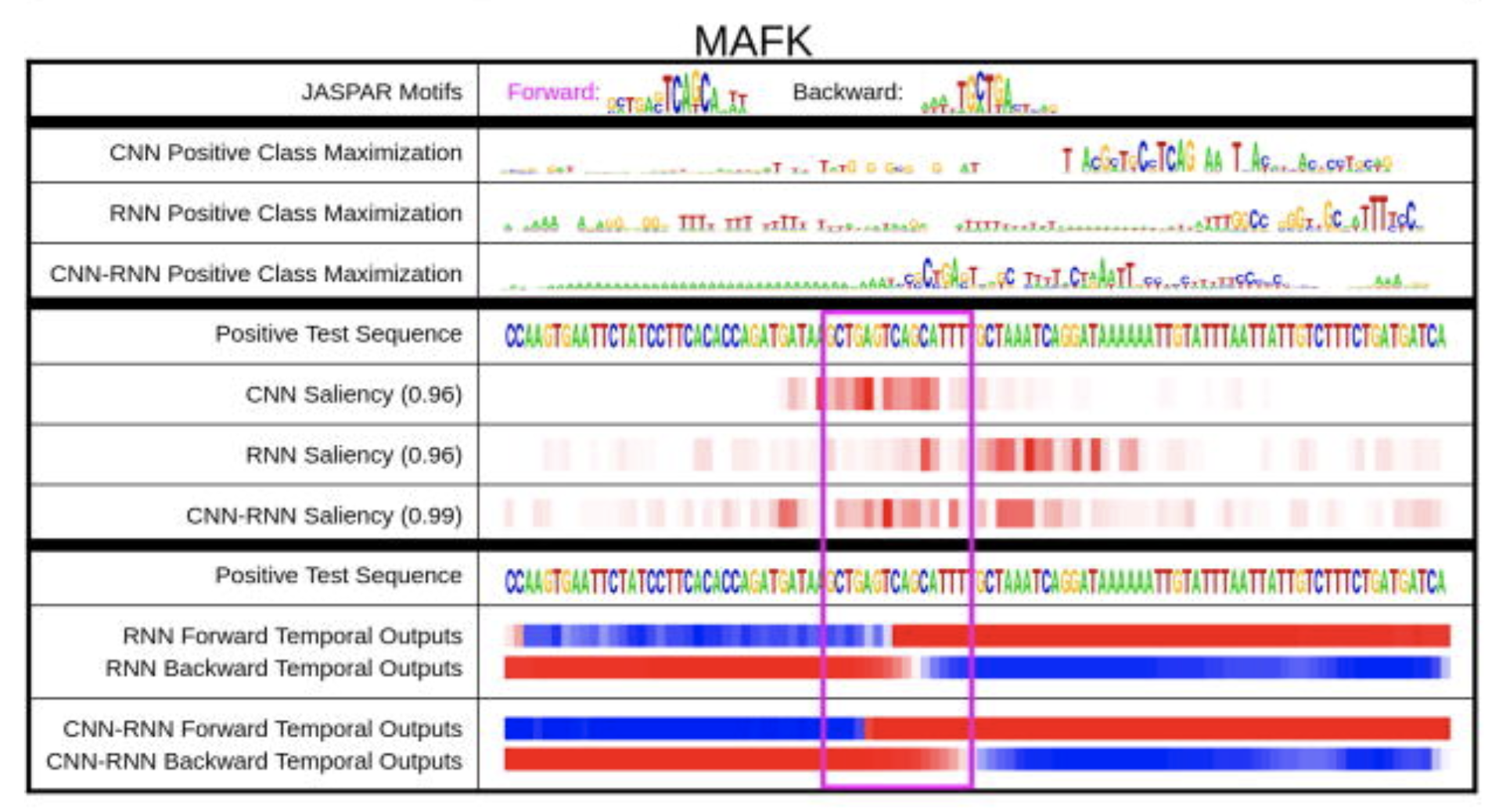 DNAPlotter
DNAPlotter EaSeq
EaSeq Edgar Synteny Plots
Edgar Synteny Plots EpiViz
EpiViz Galaxy HiCExplorer
Galaxy HiCExplorer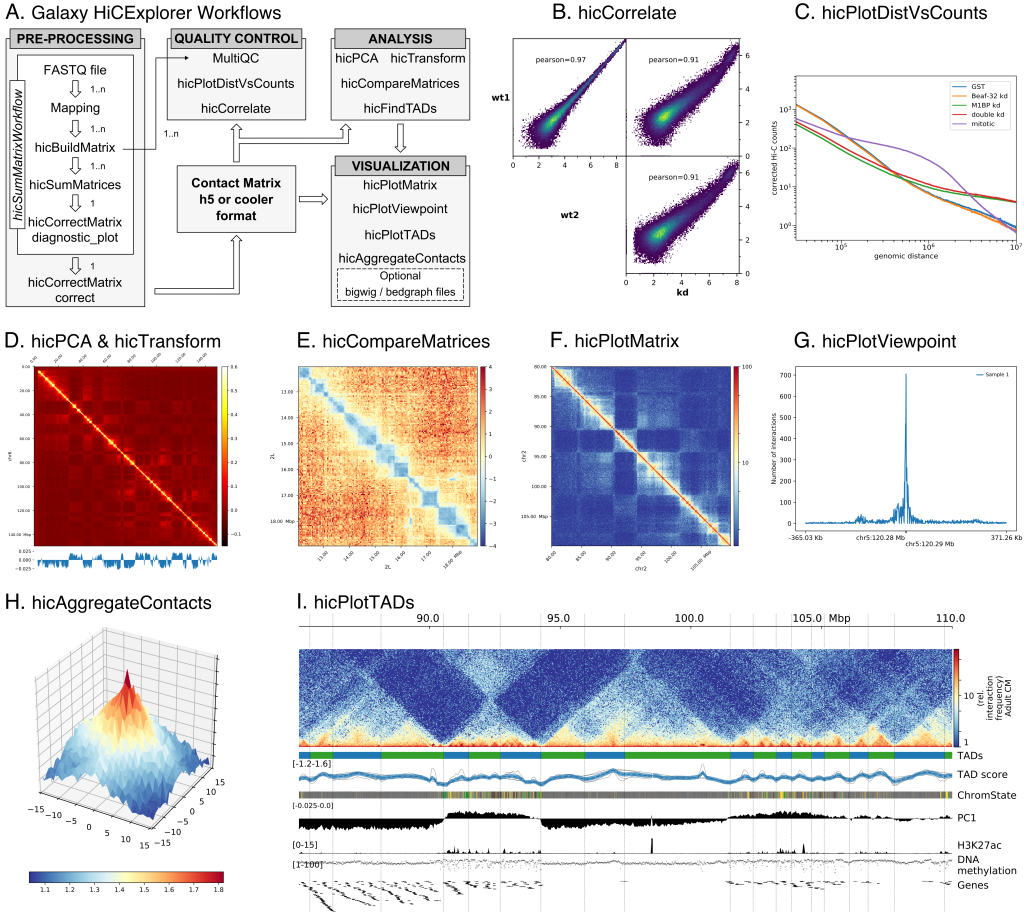 GenomeRing
GenomeRing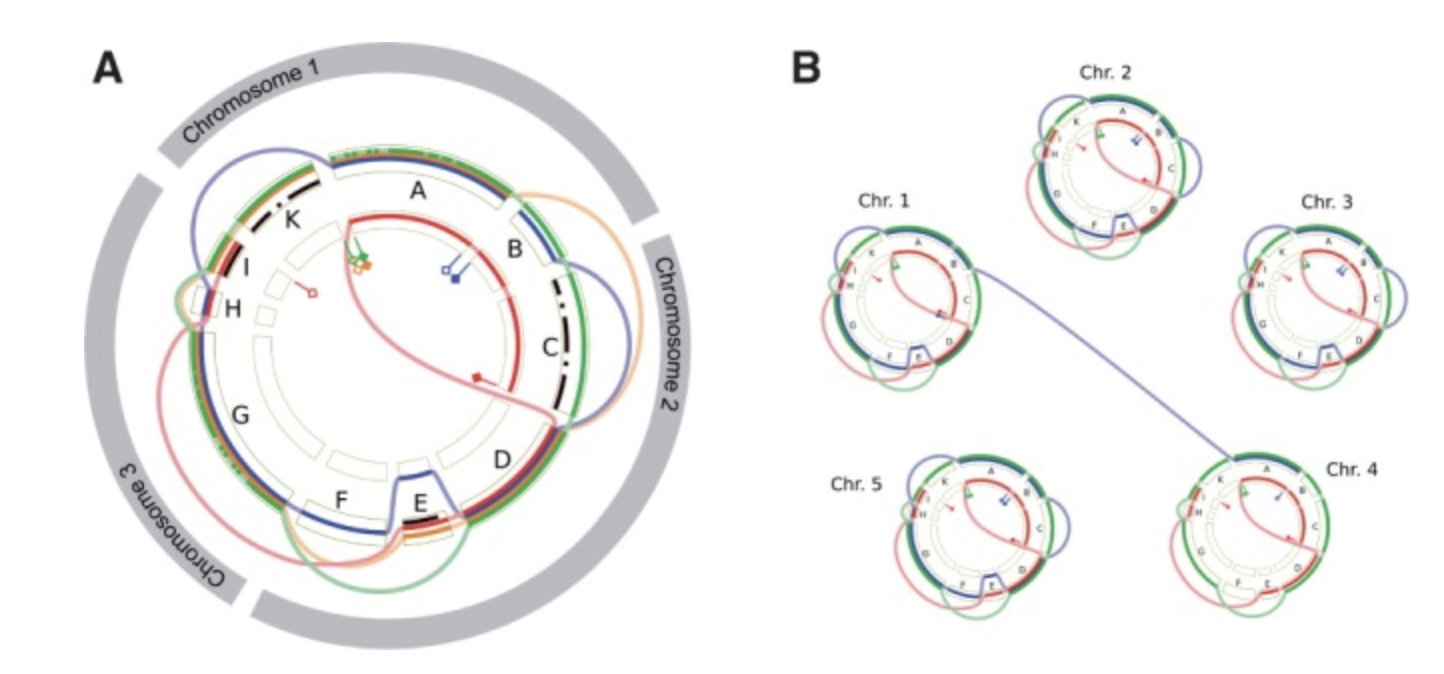 GenomeView
GenomeView genoPlotR
genoPlotR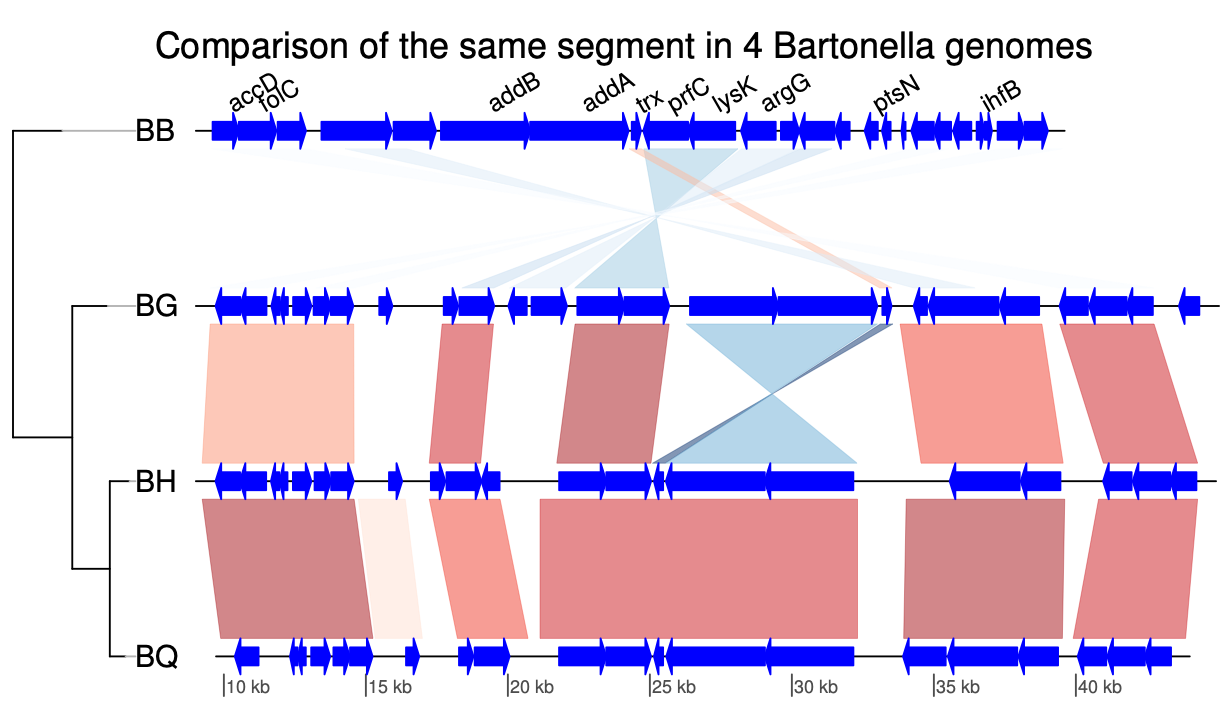 GenPlay
GenPlay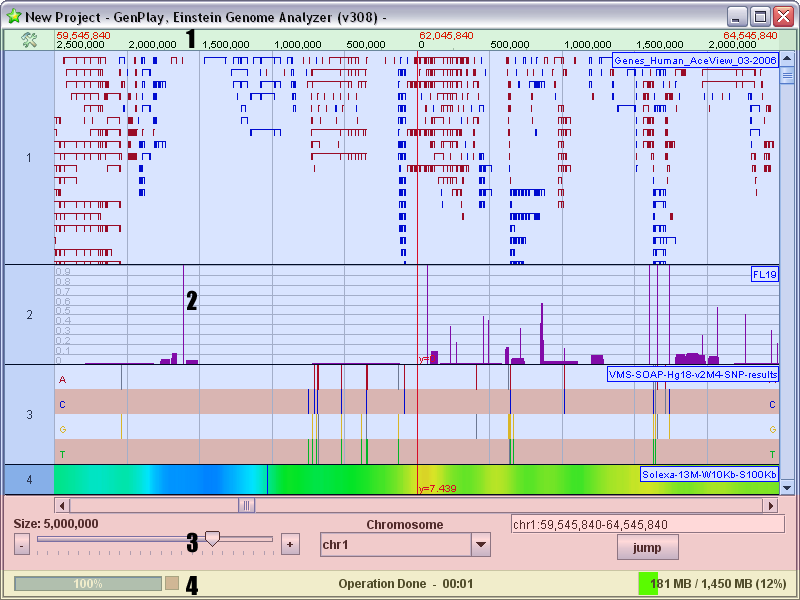 Gepard
Gepard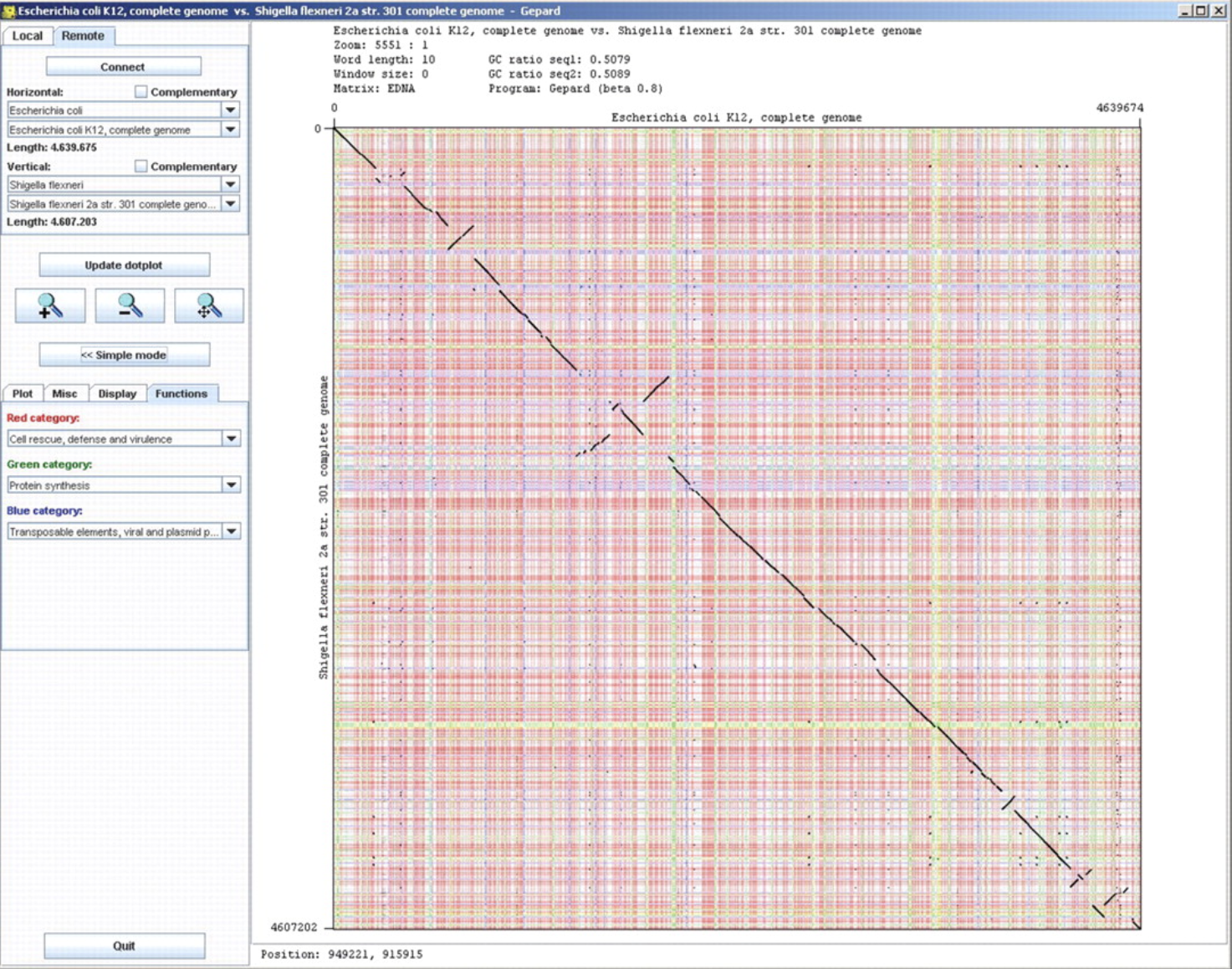 GView
GView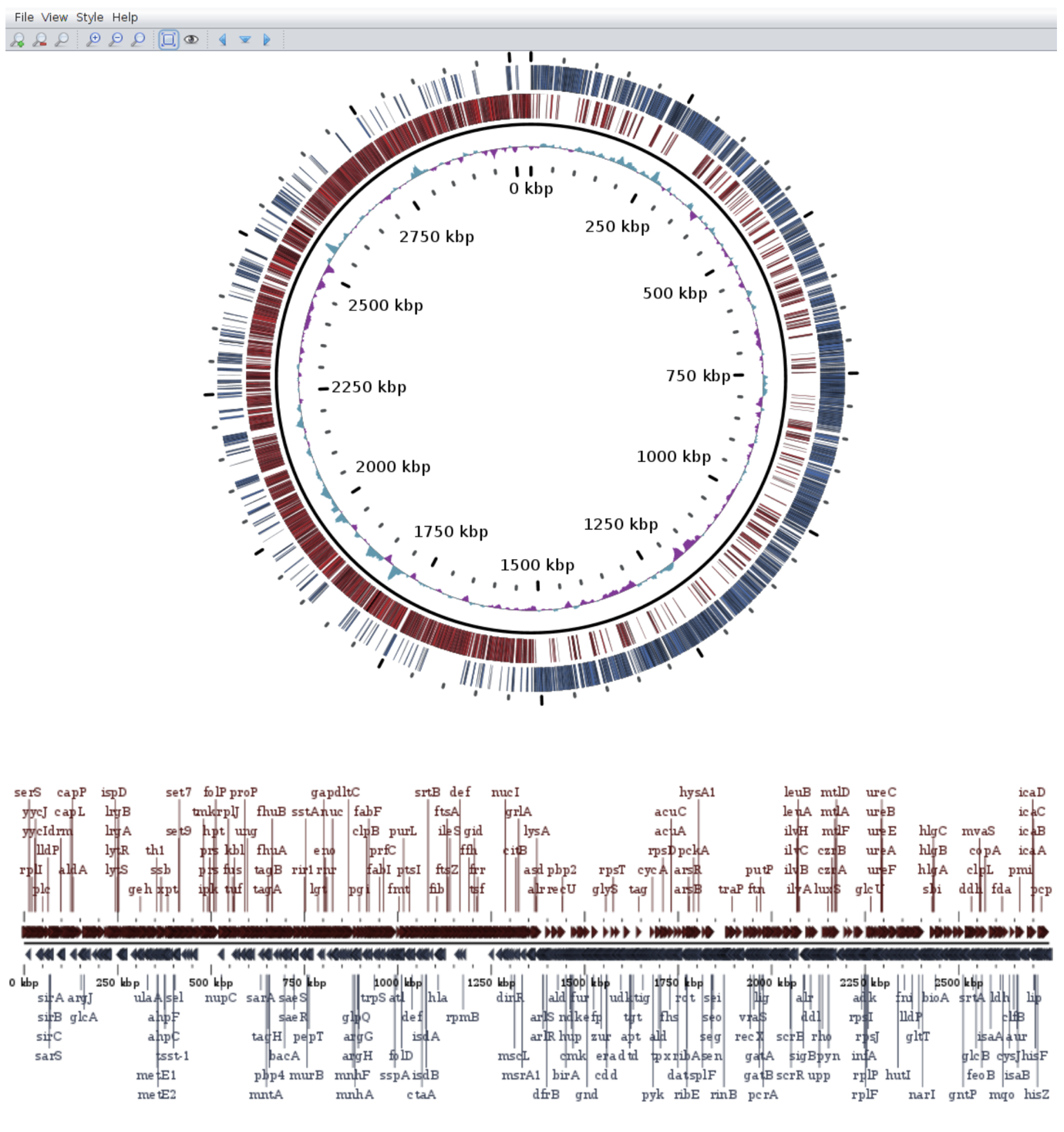 HilbertCurve
HilbertCurve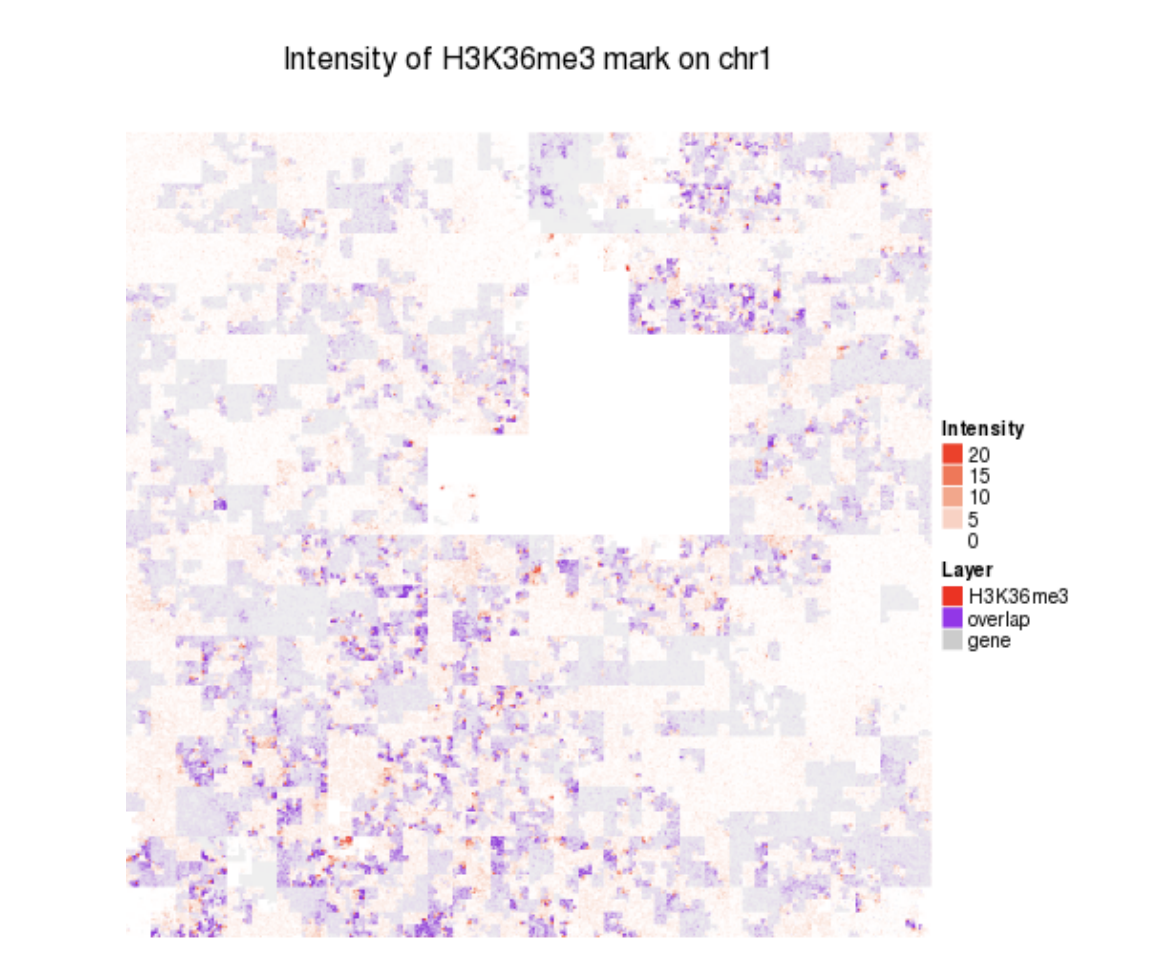 HilbertVis
HilbertVis HUGIn
HUGIn IRScope
IRScope J-Circos
J-Circos JalView
JalView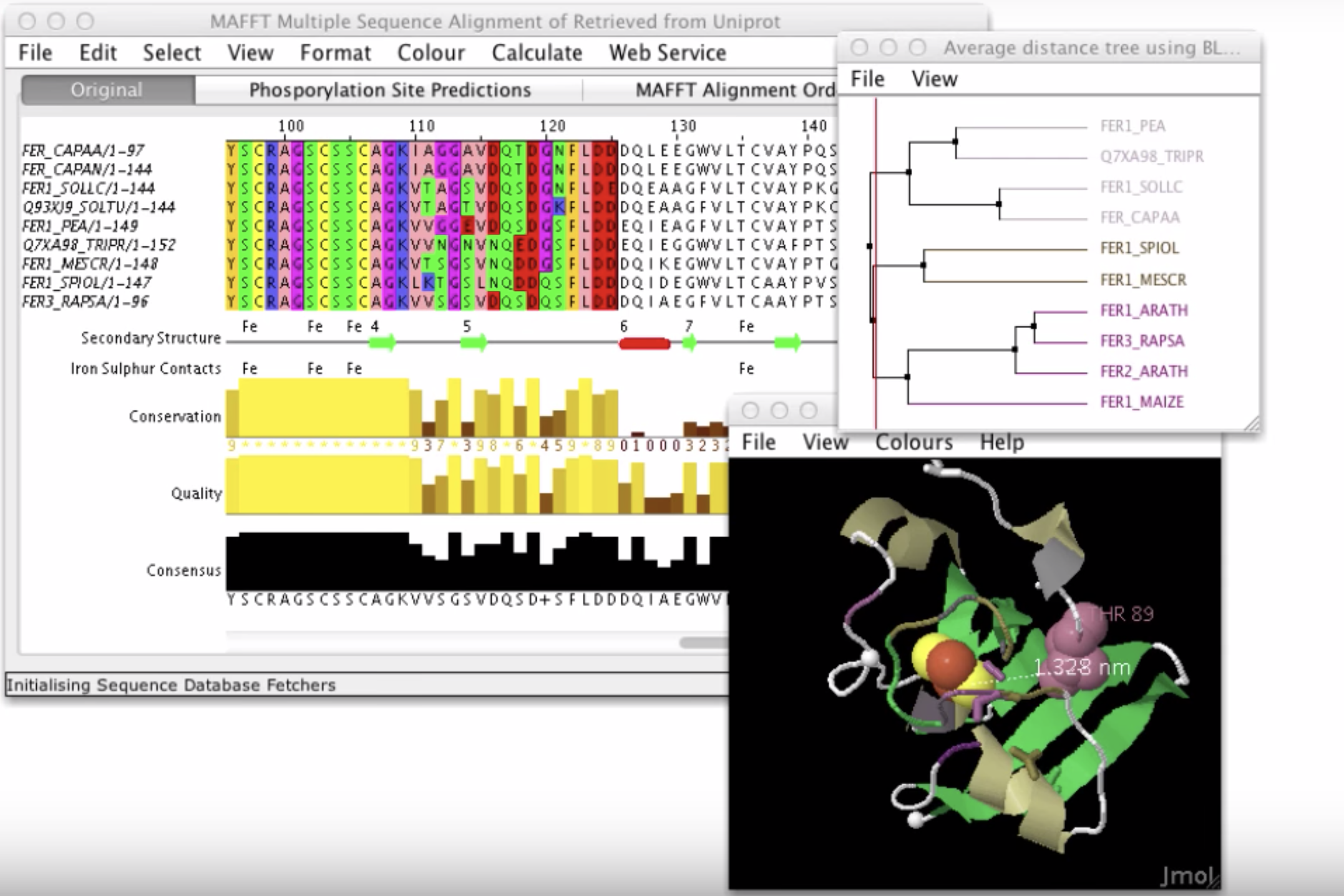 JBrowse
JBrowse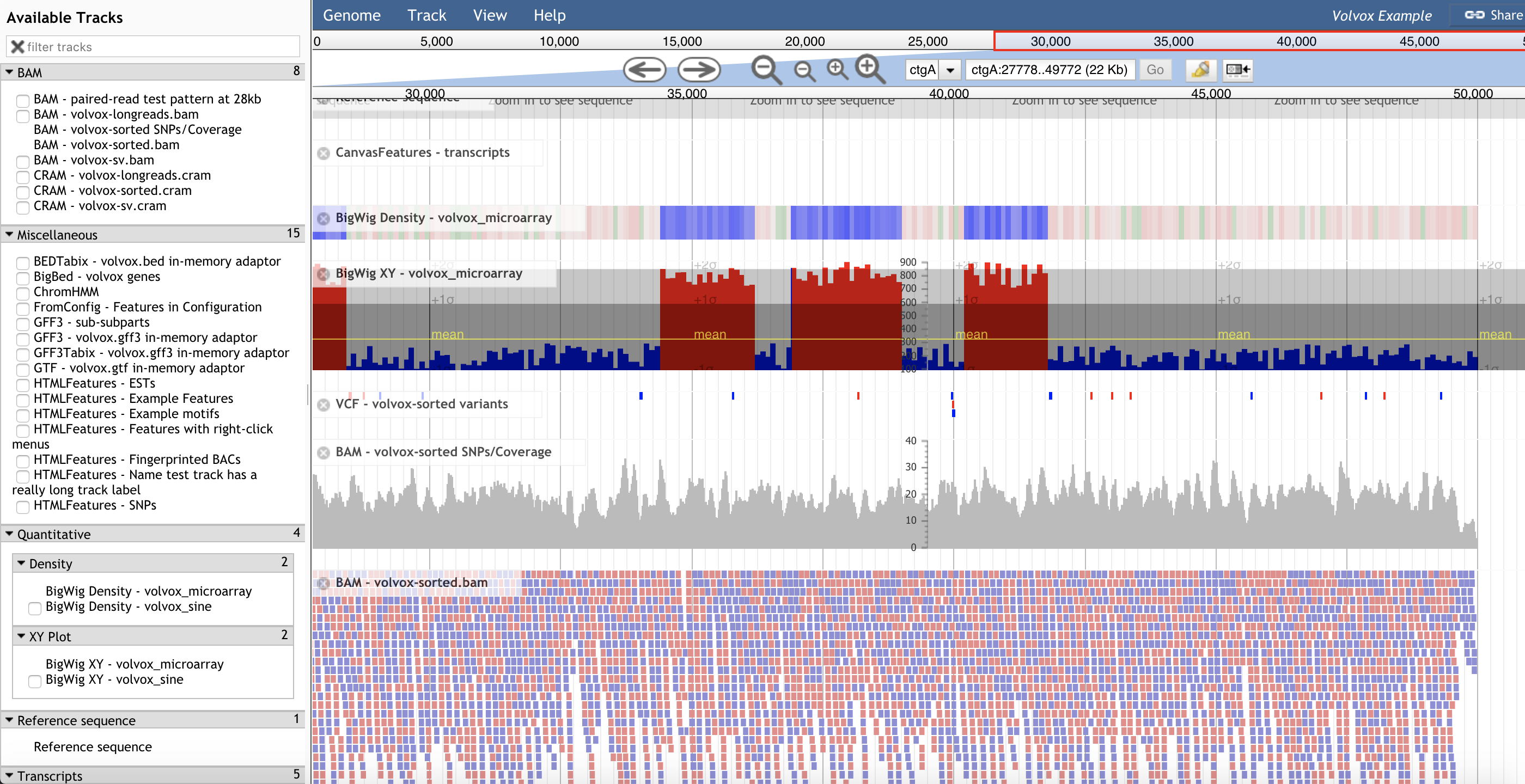 JuiceBox
JuiceBox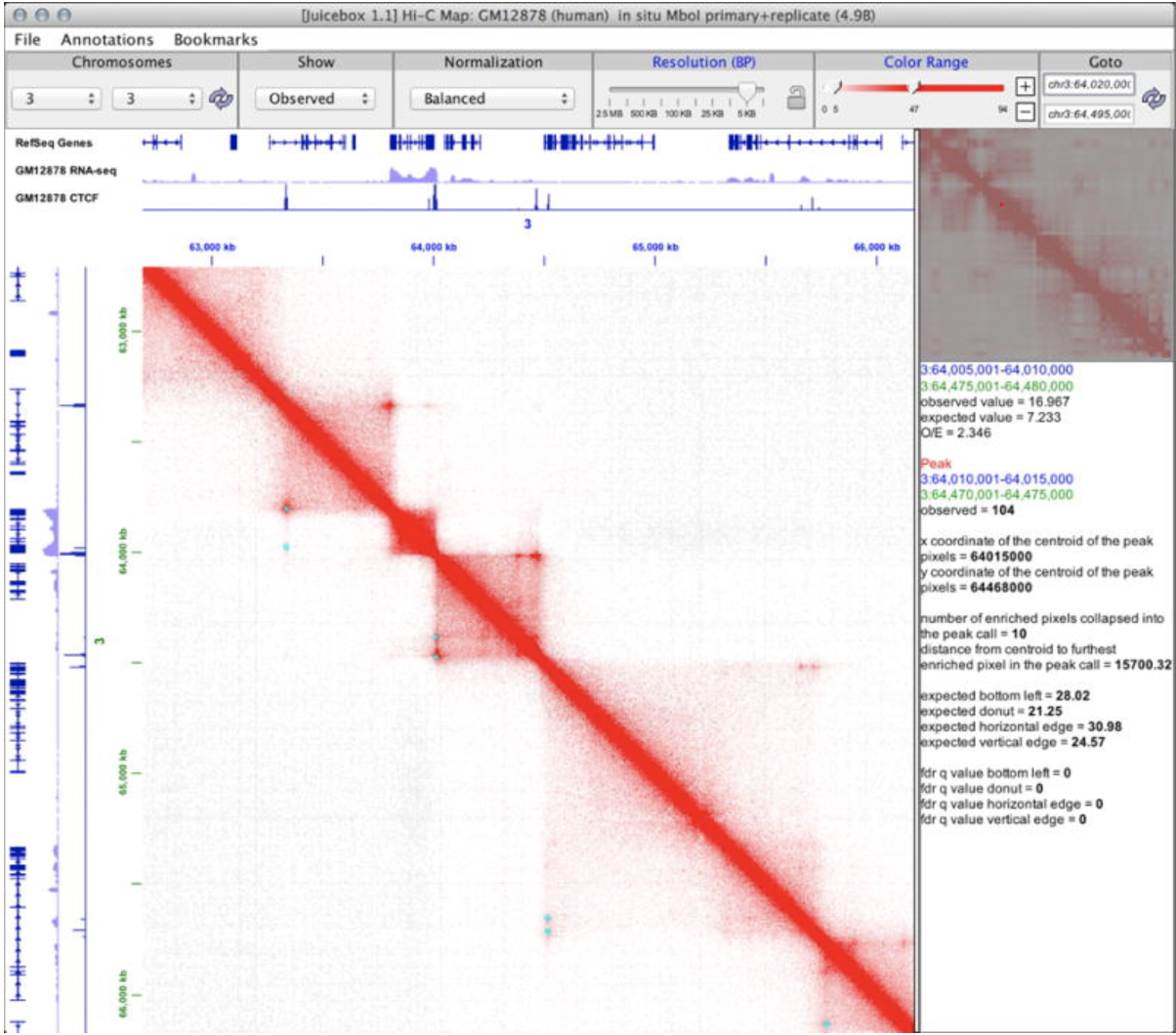 Lollipop Plot cBio
Lollipop Plot cBio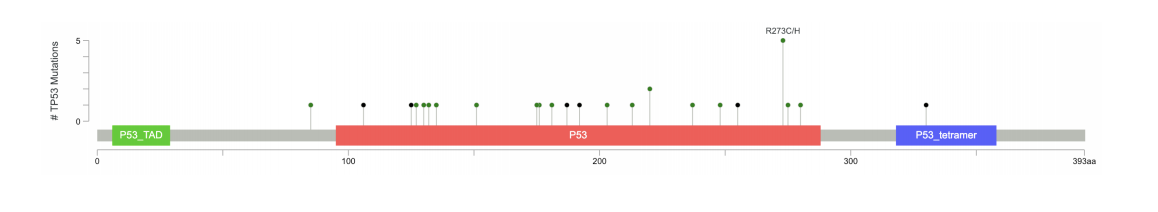 MEXPRESS
MEXPRESS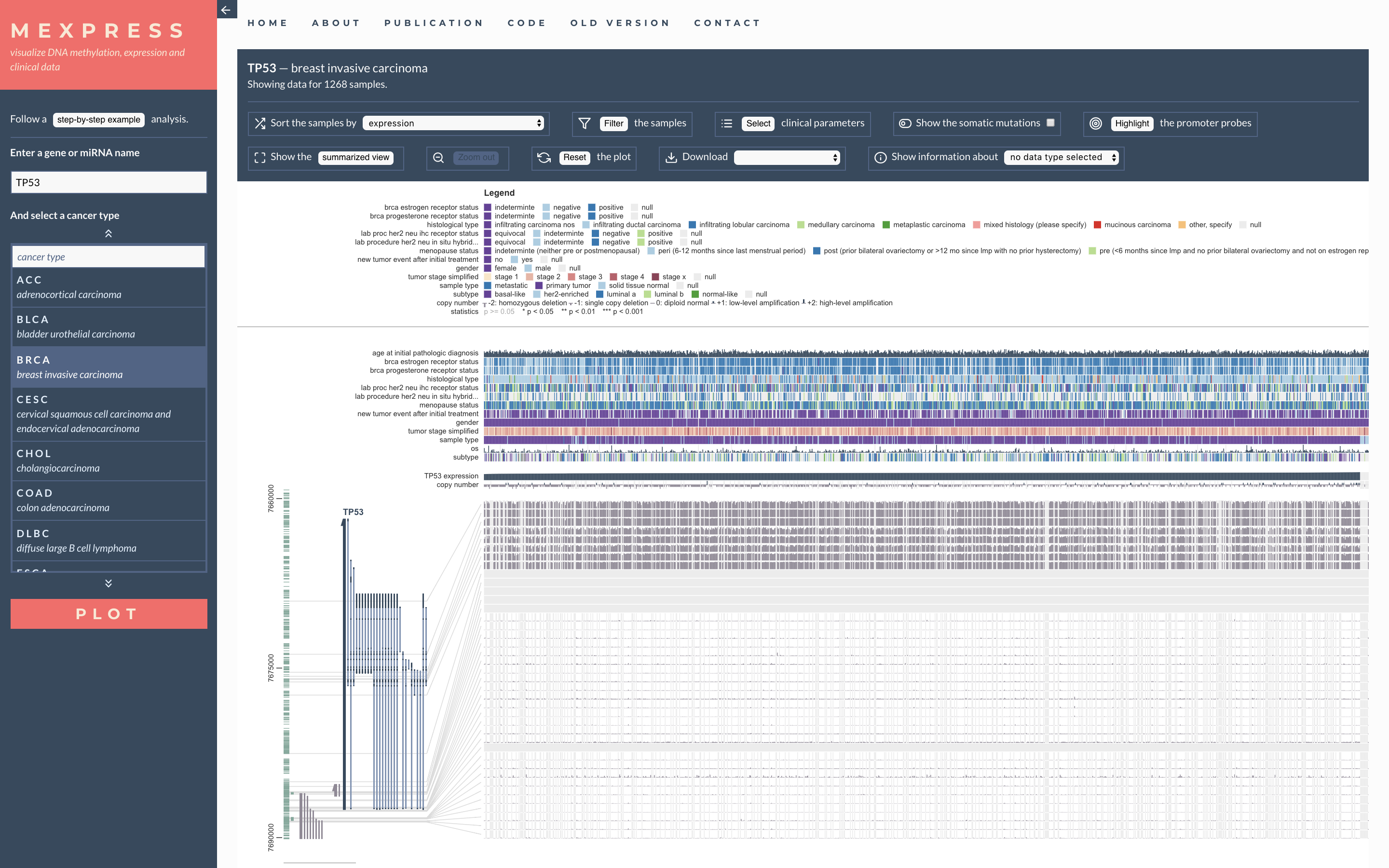 MGcV
MGcV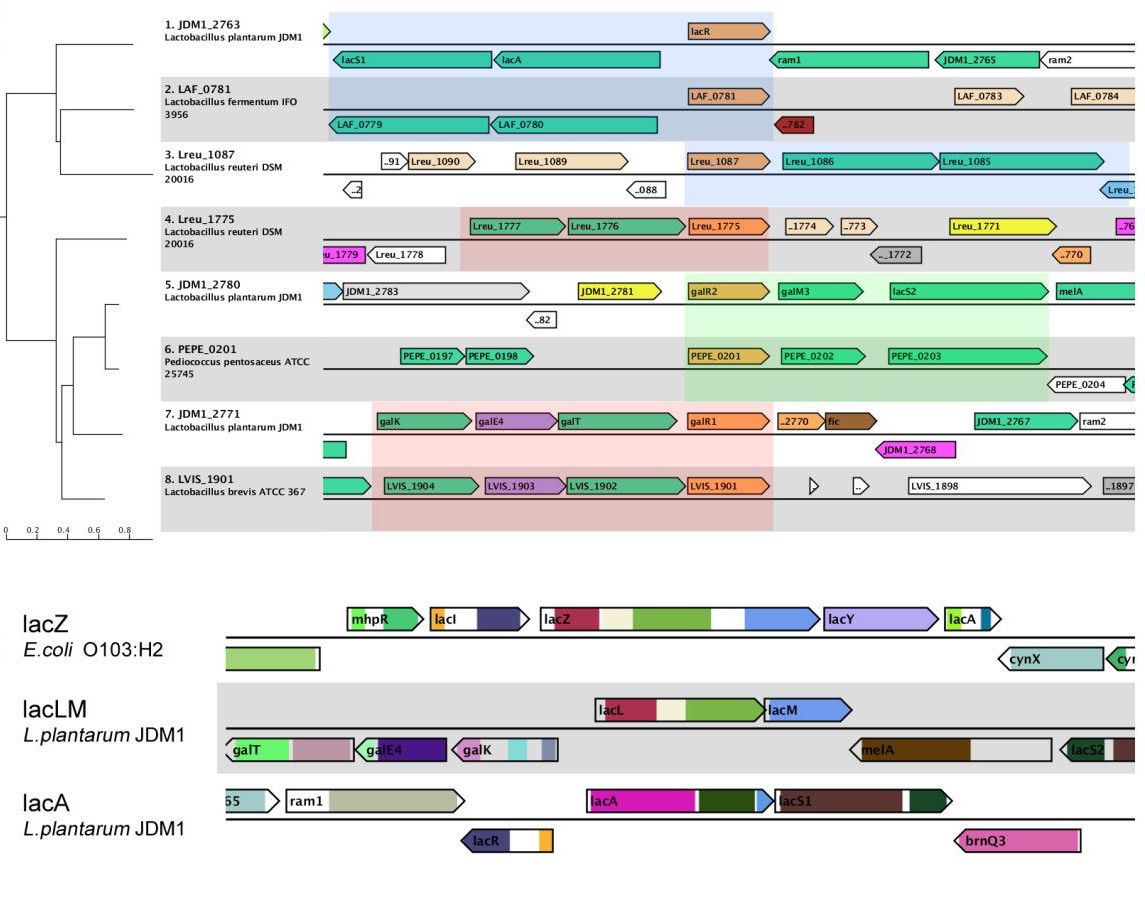 MSAViewer
MSAViewer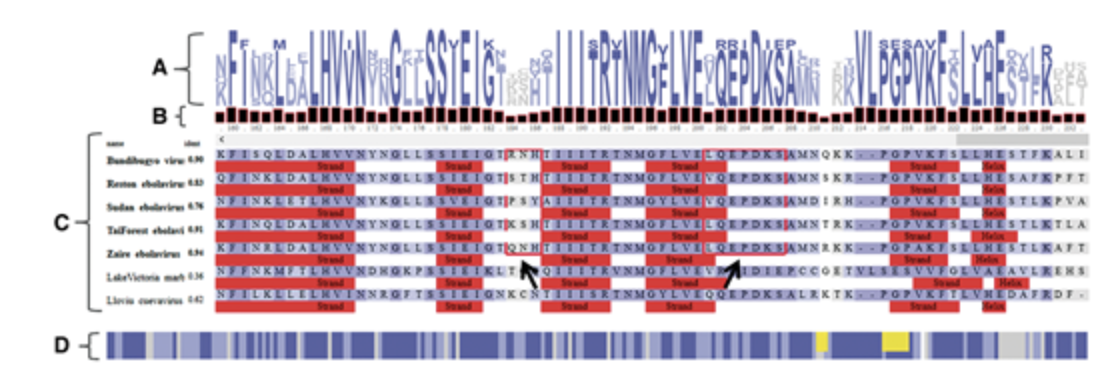 NCBI Genome Viewer
NCBI Genome Viewer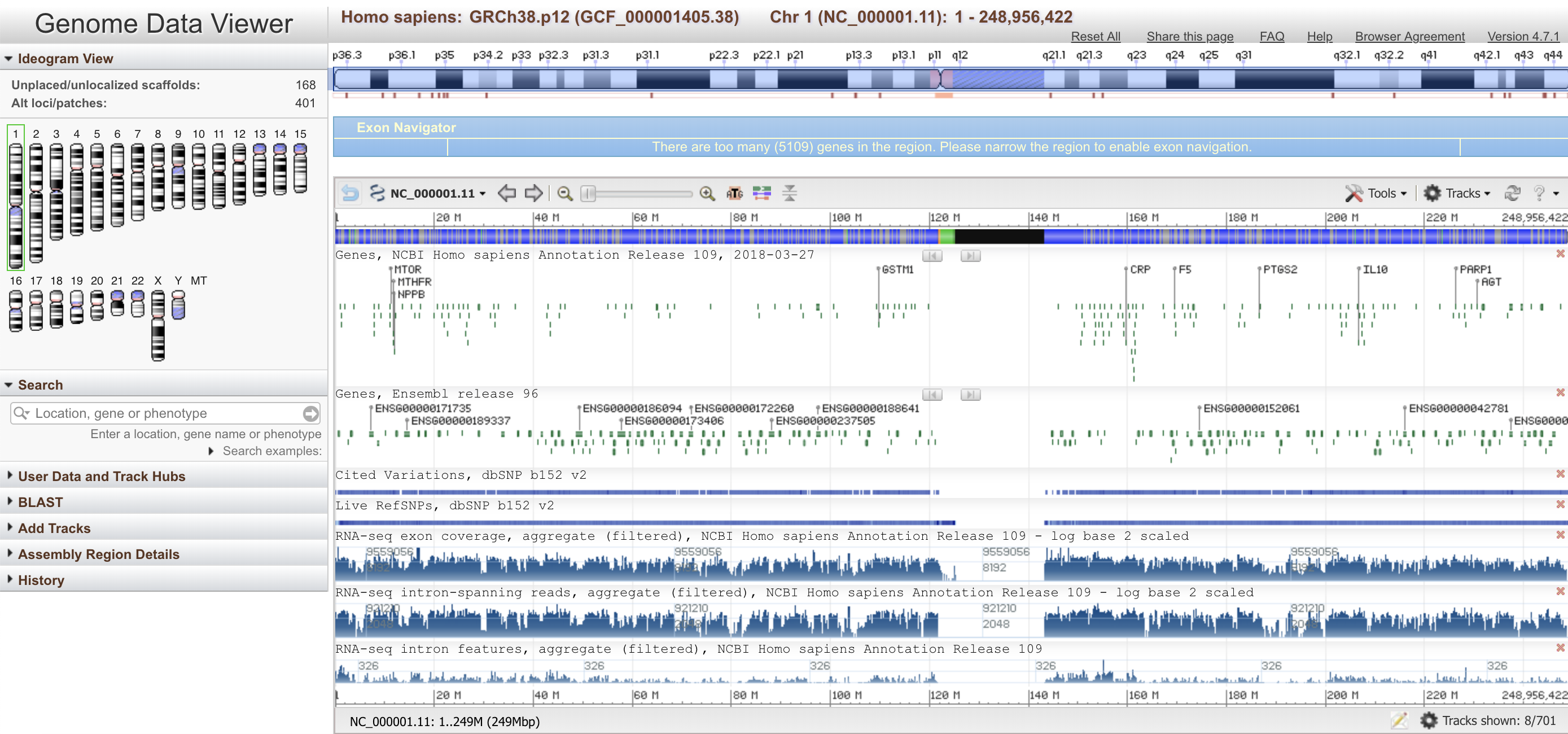 pLogo
pLogo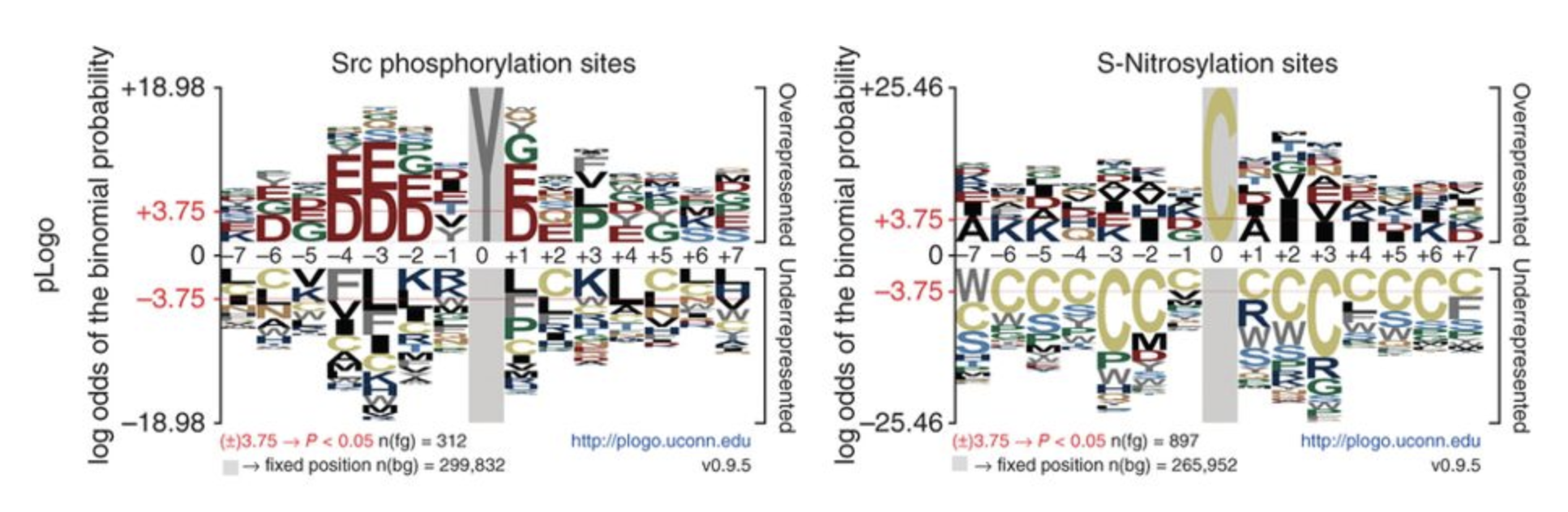 PSU 3D Genome Browser
PSU 3D Genome Browser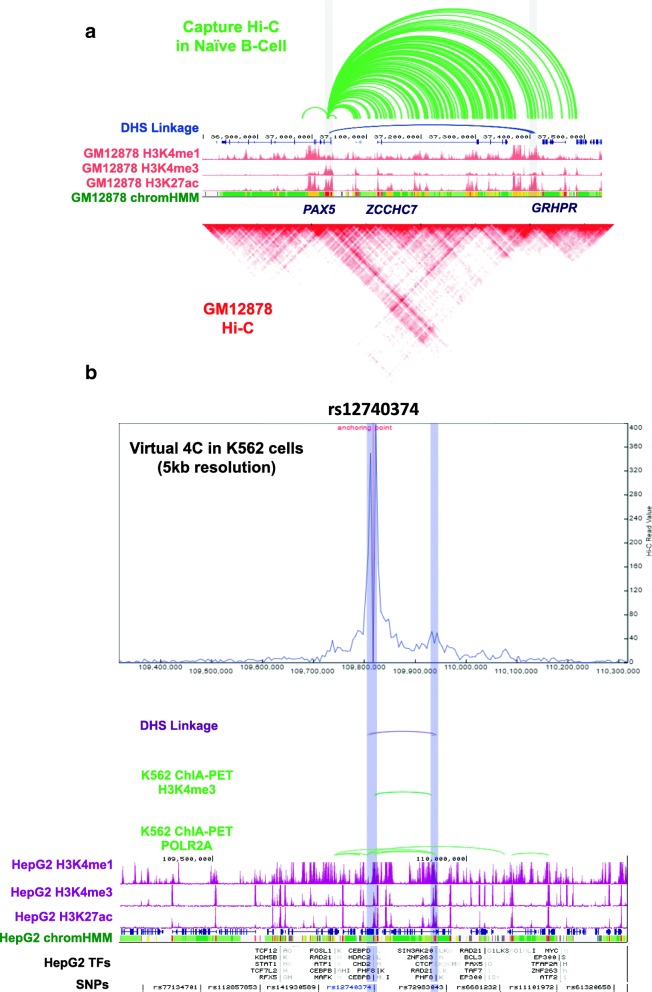 ReadXplorer
ReadXplorer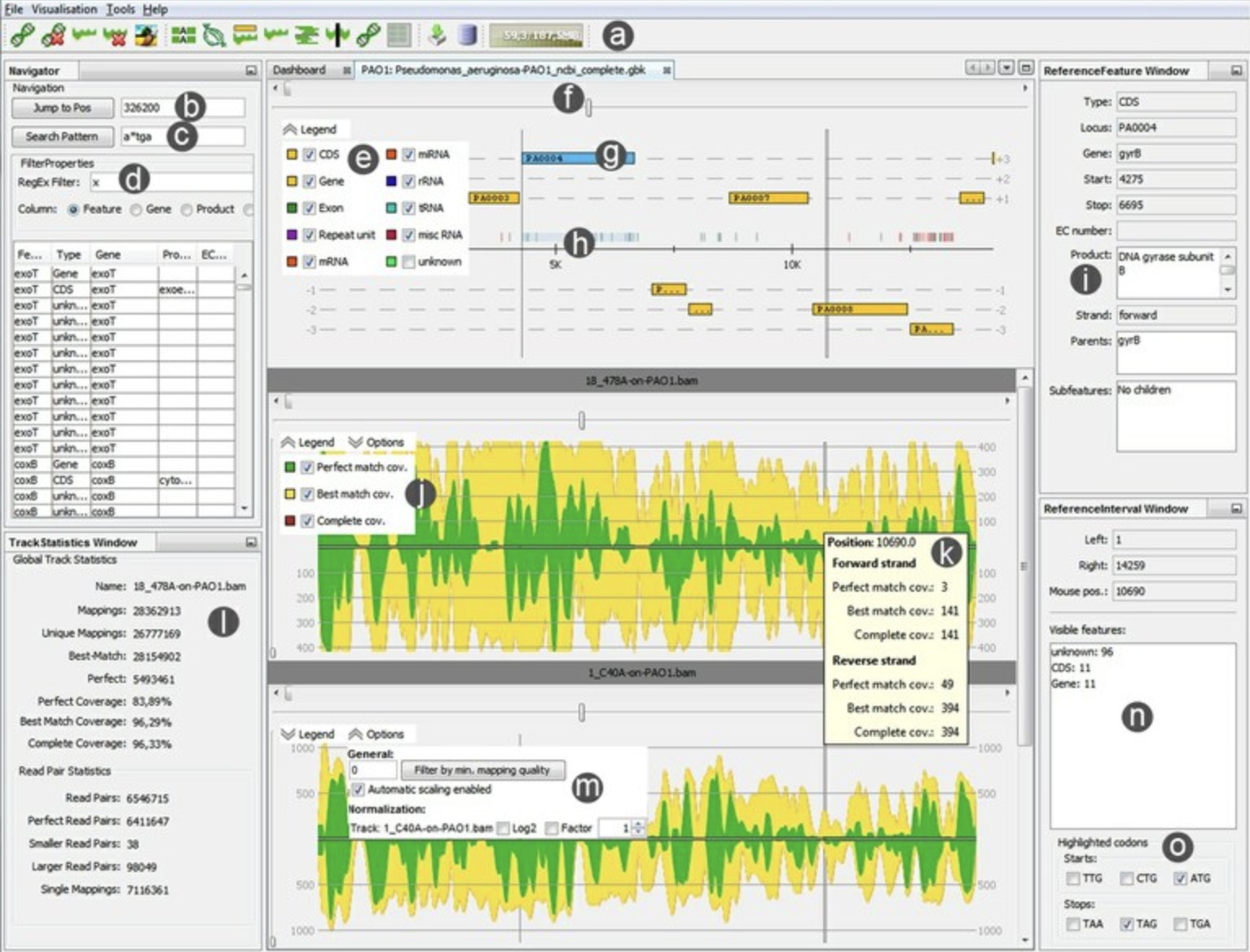 RIdeogram
RIdeogram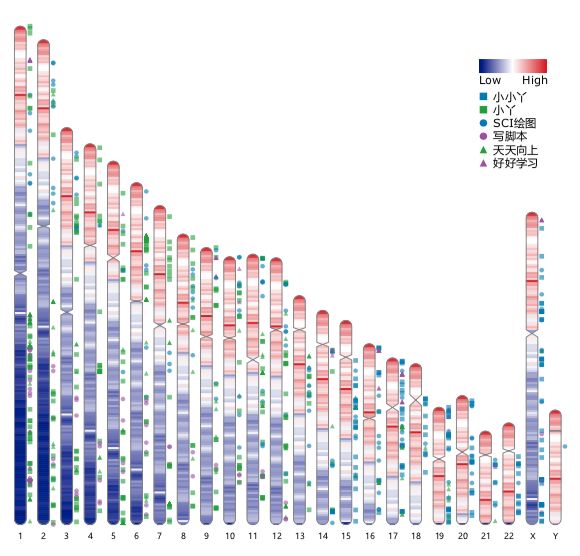 Rondo
Rondo Sashimi Plot
Sashimi Plot Sequence Bundles
Sequence Bundles Spatial DB
Spatial DB SpliceGrapher
SpliceGrapher SplicePlot
SplicePlot SpliceSeq
SpliceSeq svist4get
svist4get SynMap2
SynMap2 SynteBase and SynteView
SynteBase and SynteView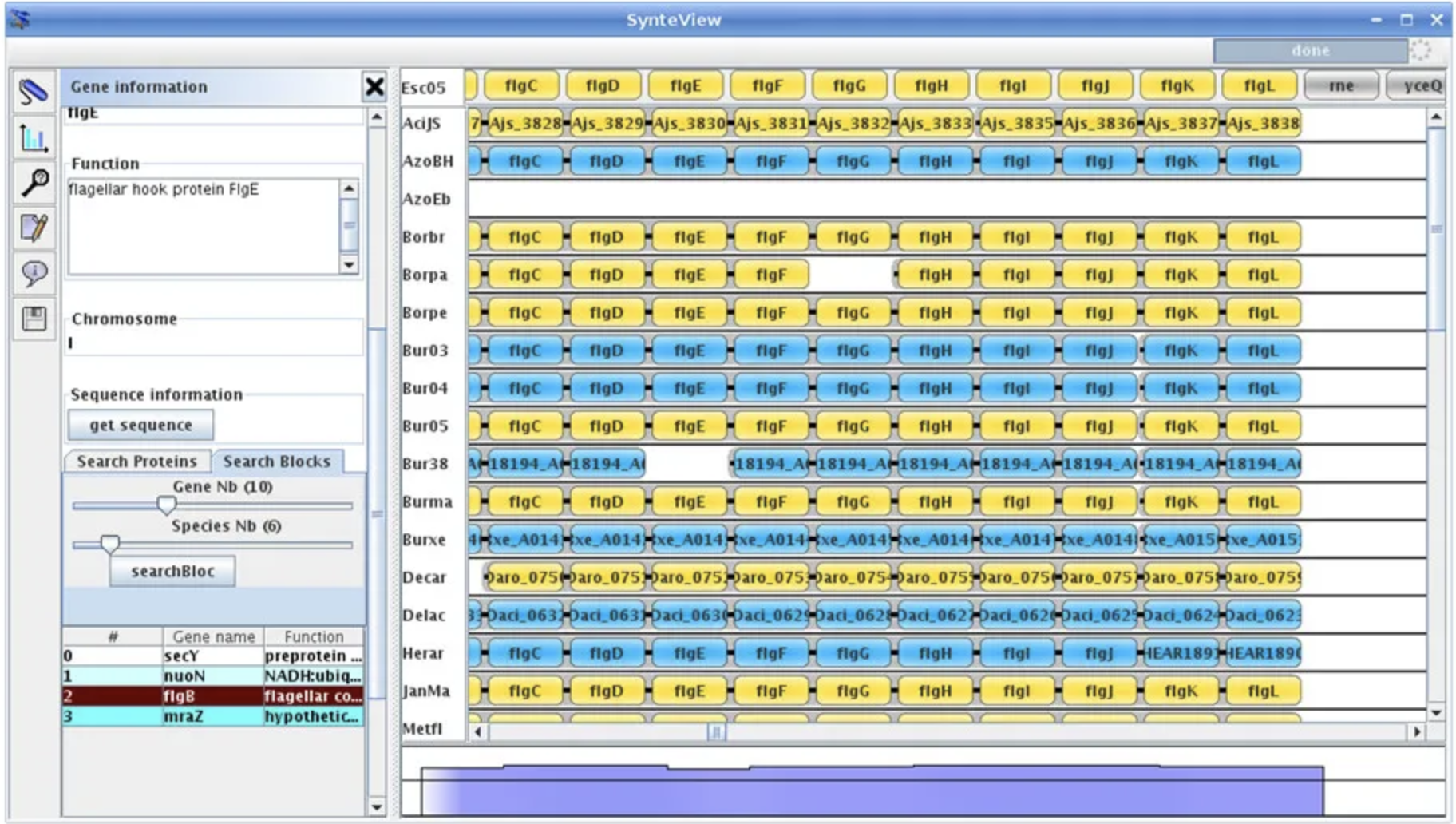 TFmotifView
TFmotifView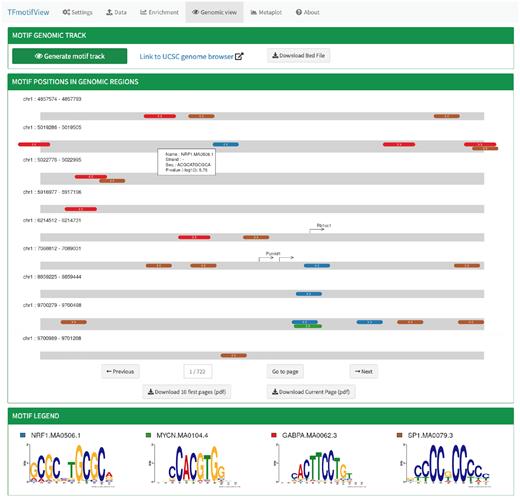 Two Sample Logo
Two Sample Logo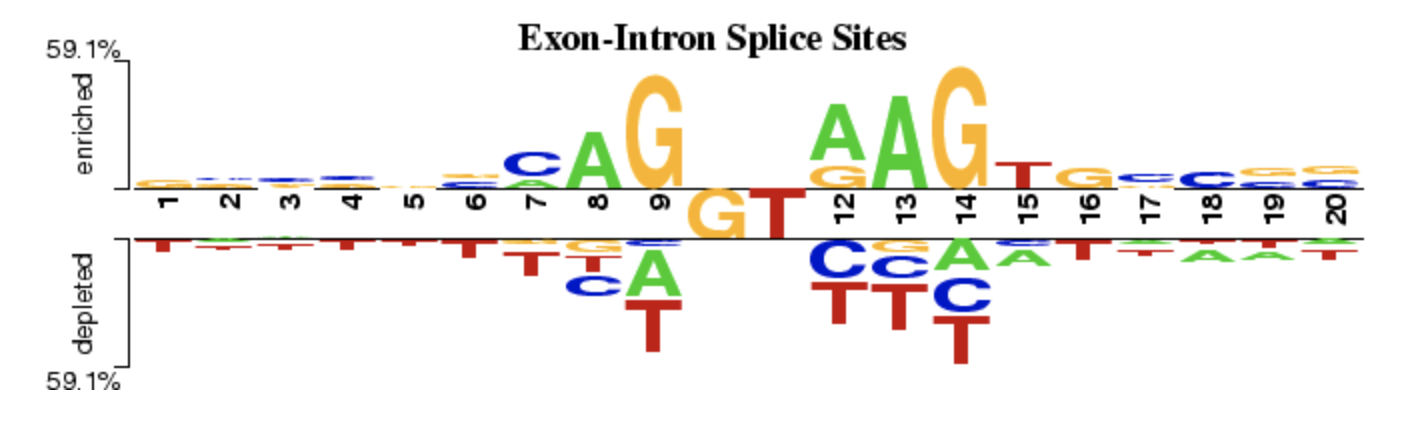 UCSC Genome Browser
UCSC Genome Browser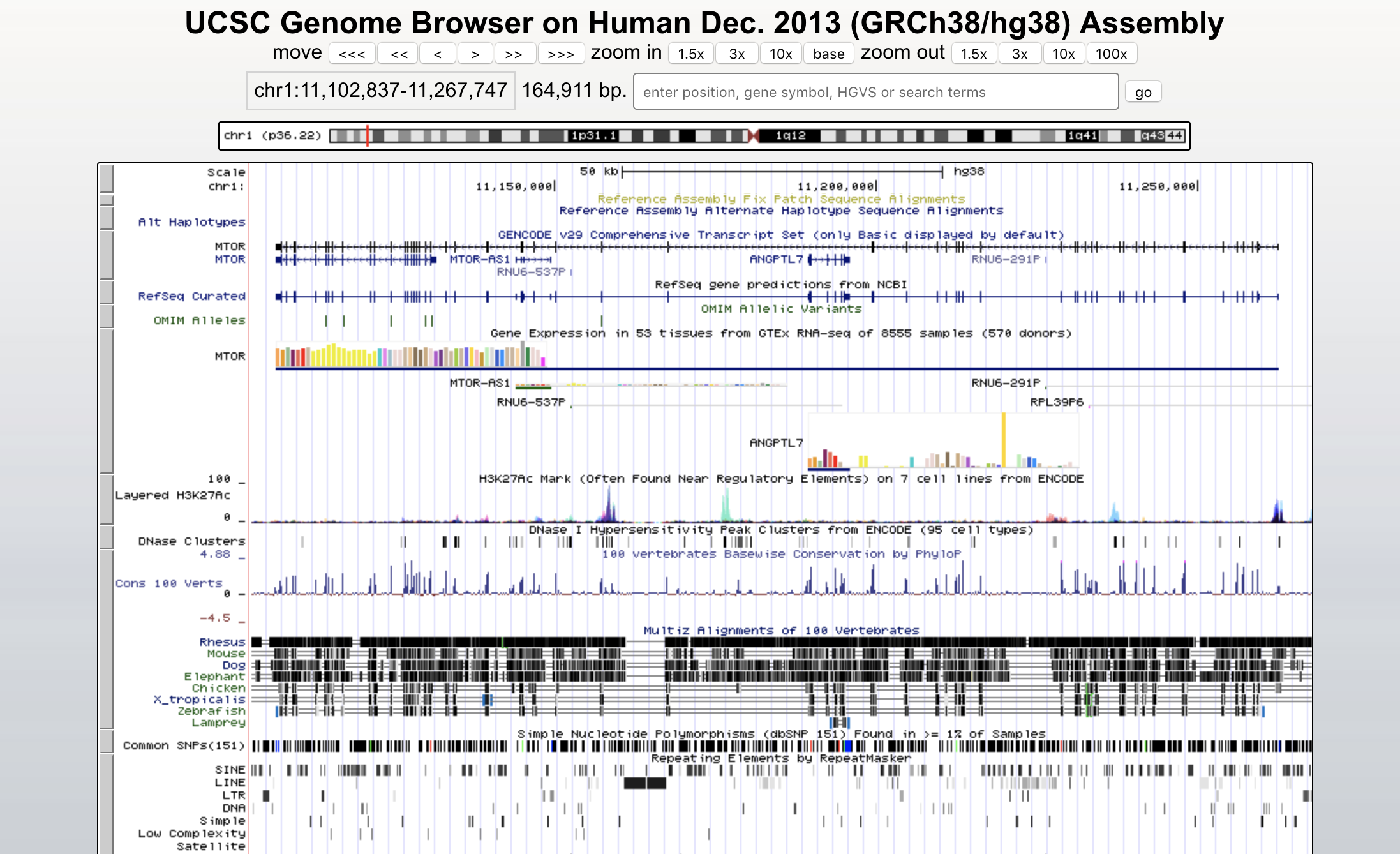 Variant View
Variant View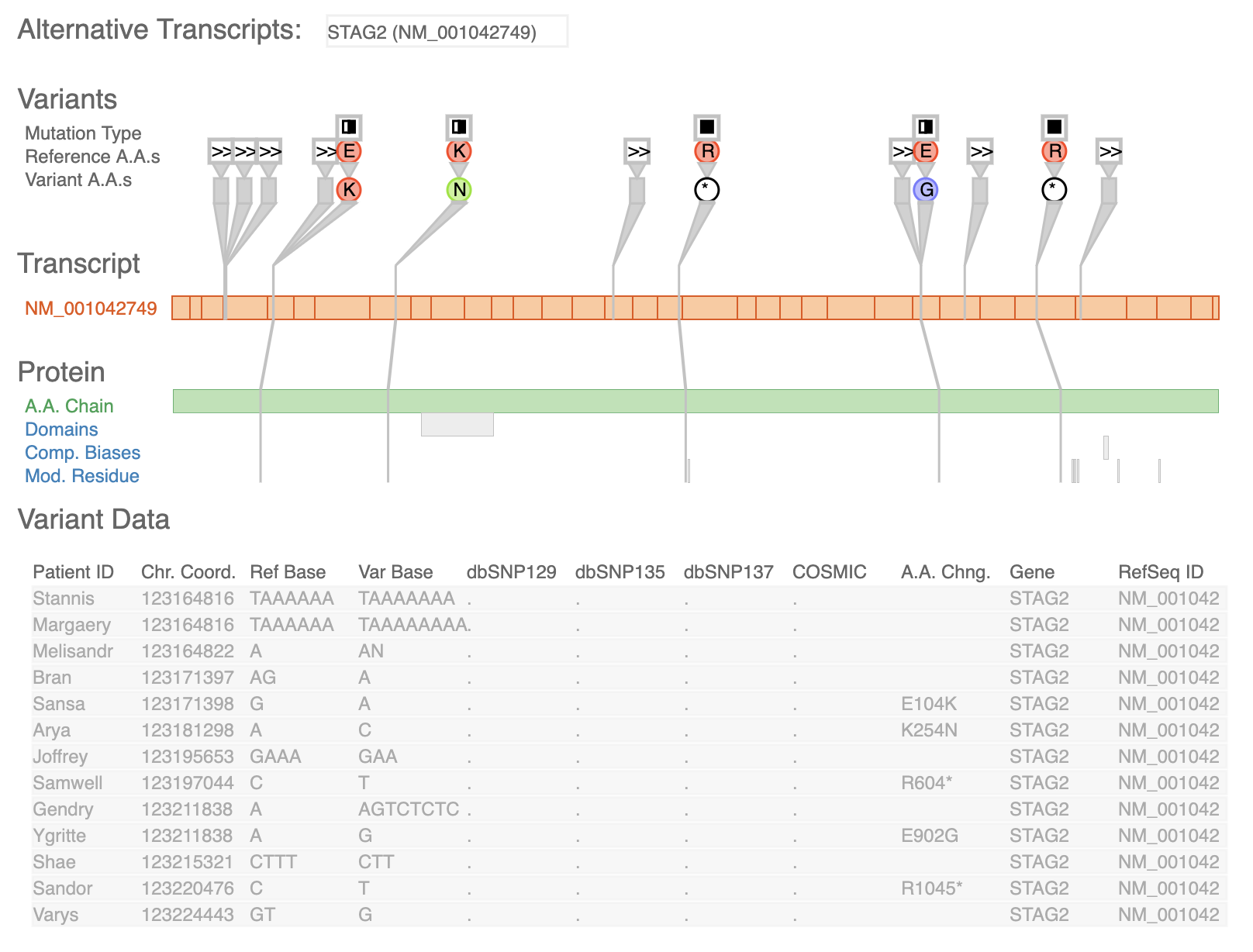 VCF Plotein
VCF Plotein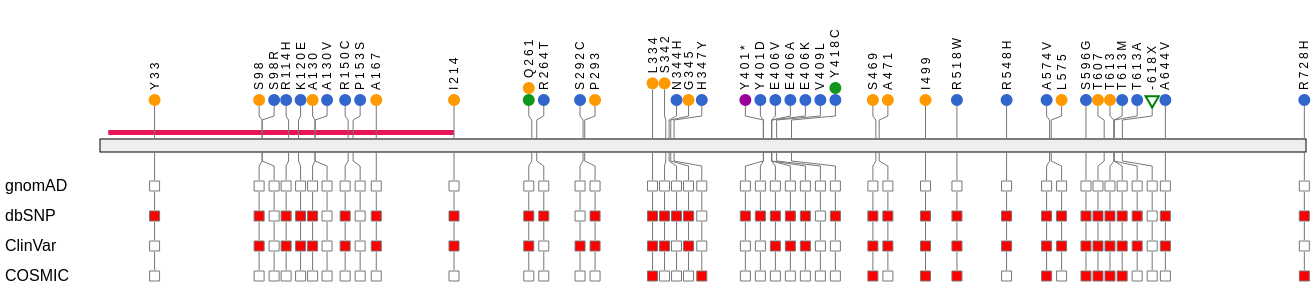 Vials
Vials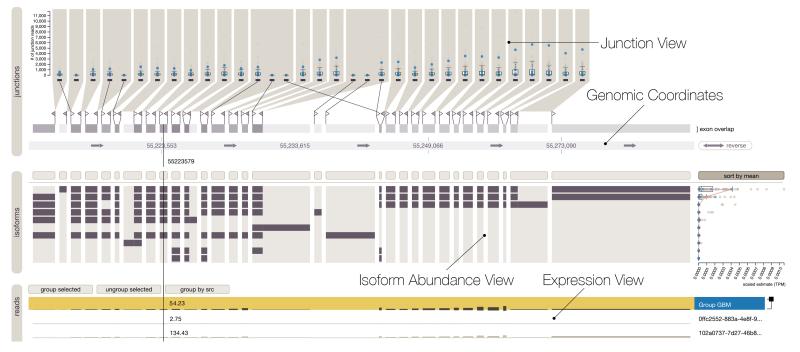 Vista Dot
Vista Dot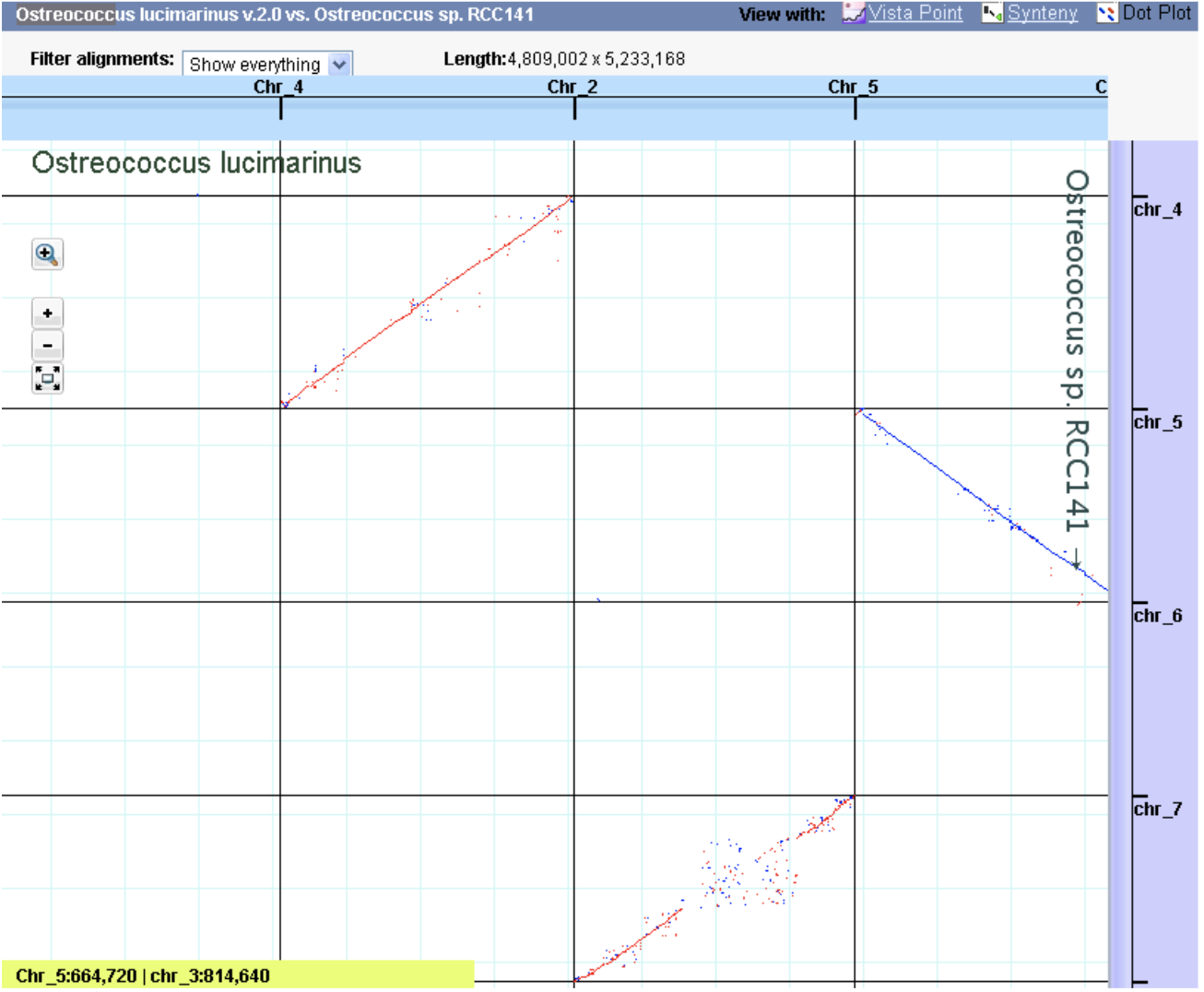 VistaPoint
VistaPoint WashU Epigenome Browser
WashU Epigenome Browser WebLogo
WebLogo
 3dViewer
3dViewer AliView
AliView Alvis
Alvis BactoGenie
BactoGenie CGView
CGView CGView Server
CGView Server Cinteny
Cinteny Circos
Circos Civi
Civi ClicO Free Service
ClicO Free Service CNVkit
CNVkit Combo
Combo CRISPResso2
CRISPResso2 Dalliance
Dalliance Deep Motif Dashboard
Deep Motif Dashboard DNAPlotter
DNAPlotter EaSeq
EaSeq Edgar Synteny Plots
Edgar Synteny Plots EpiViz
EpiViz Galaxy HiCExplorer
Galaxy HiCExplorer GenomeRing
GenomeRing GenomeView
GenomeView genoPlotR
genoPlotR GenPlay
GenPlay Gepard
Gepard GView
GView HilbertCurve
HilbertCurve HilbertVis
HilbertVis HUGIn
HUGIn IRScope
IRScope J-Circos
J-Circos JalView
JalView JBrowse
JBrowse JuiceBox
JuiceBox Lollipop Plot cBio
Lollipop Plot cBio MEXPRESS
MEXPRESS MGcV
MGcV MSAViewer
MSAViewer NCBI Genome Viewer
NCBI Genome Viewer pLogo
pLogo PSU 3D Genome Browser
PSU 3D Genome Browser ReadXplorer
ReadXplorer RIdeogram
RIdeogram Rondo
Rondo Sashimi Plot
Sashimi Plot Sequence Bundles
Sequence Bundles Spatial DB
Spatial DB SpliceGrapher
SpliceGrapher SplicePlot
SplicePlot SpliceSeq
SpliceSeq svist4get
svist4get SynMap2
SynMap2 SynteBase and SynteView
SynteBase and SynteView TFmotifView
TFmotifView Two Sample Logo
Two Sample Logo UCSC Genome Browser
UCSC Genome Browser Variant View
Variant View VCF Plotein
VCF Plotein Vials
Vials Vista Dot
Vista Dot VistaPoint
VistaPoint WashU Epigenome Browser
WashU Epigenome Browser WebLogo
WebLogo
Multiple Views

Multiple views allow for visualizations with multiple scales and foci. Multiple views can be (i) independent, (ii) weakly linked, (iii) medium linked or (iv) strongly linked. While independent views are not connected in any way, weakly linked views are linked by brushing and linking. Medium linked views share navigation, for example two views are at different scales, but zooming always affects both. Strongly linked views share genomic coordinates at one axis and can also share tracks that are aligned to the axis.
ABrowse Apollo
Apollo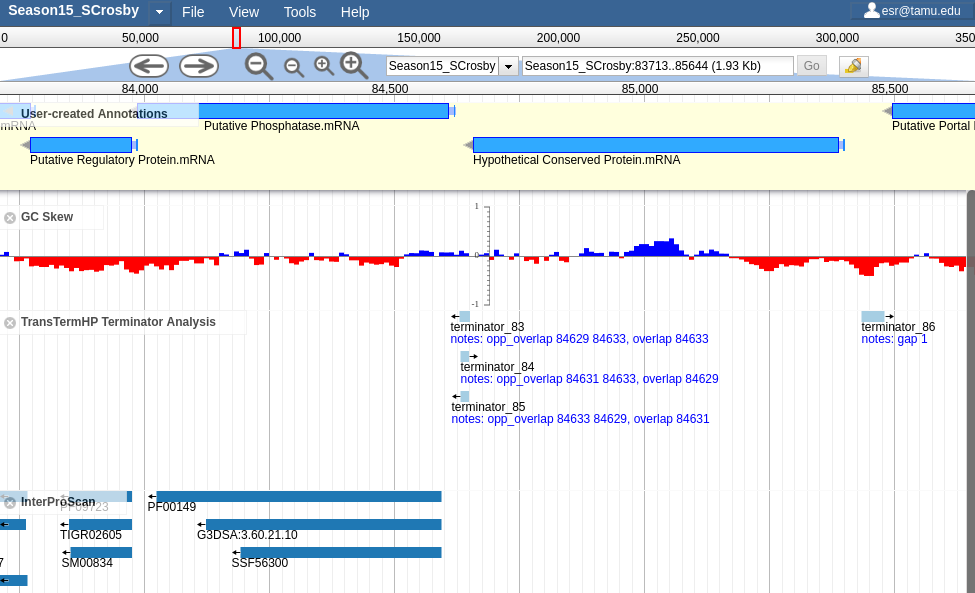 BedSect
BedSect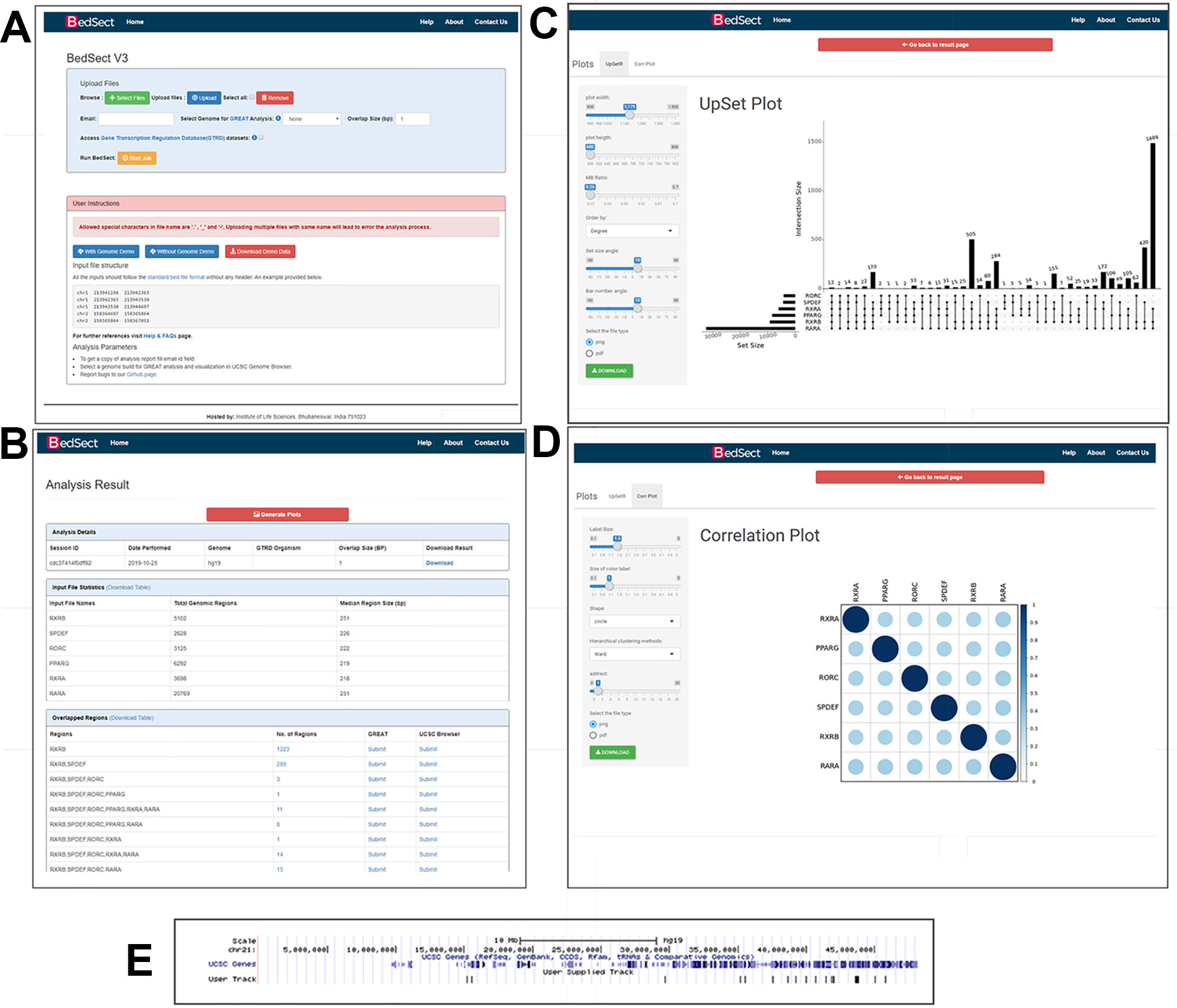 CEpBrowser
CEpBrowser Cooler
Cooler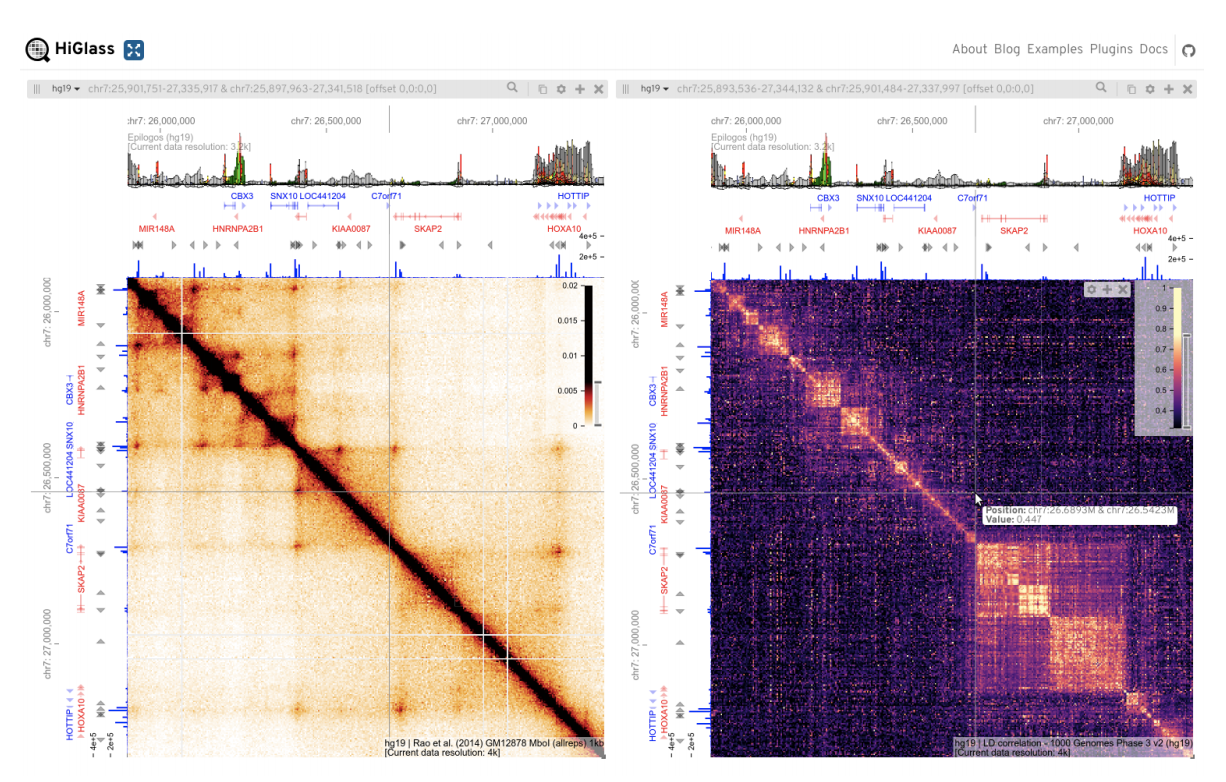 CRAMER
CRAMER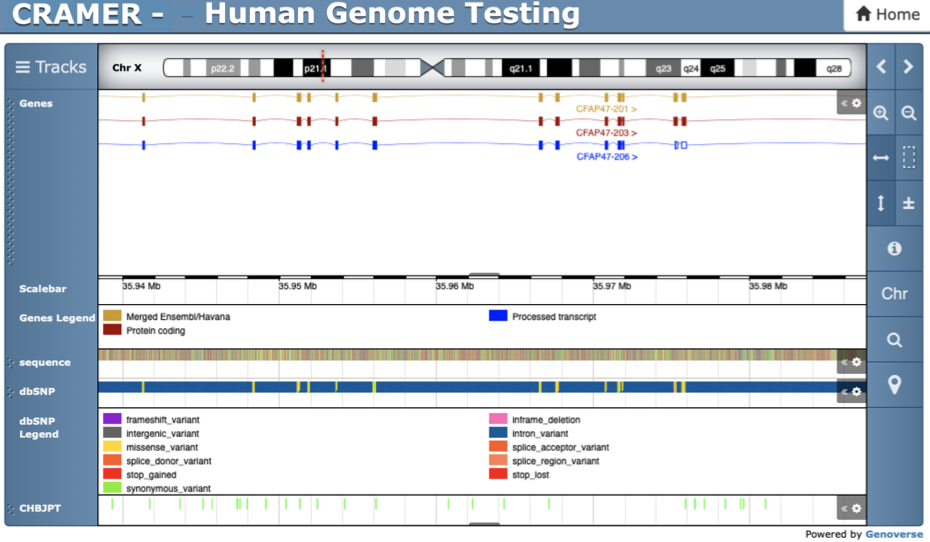 deepTools Heatmap
deepTools Heatmap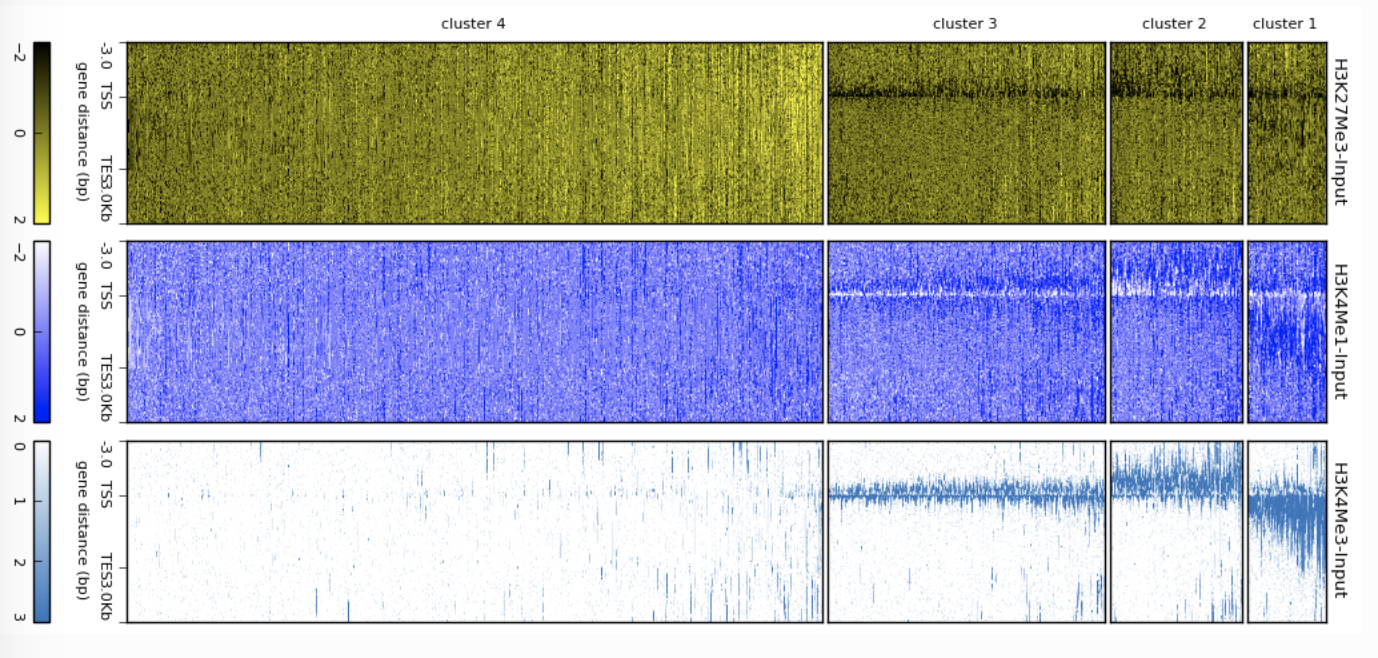 Edgar Genome Browser
Edgar Genome Browser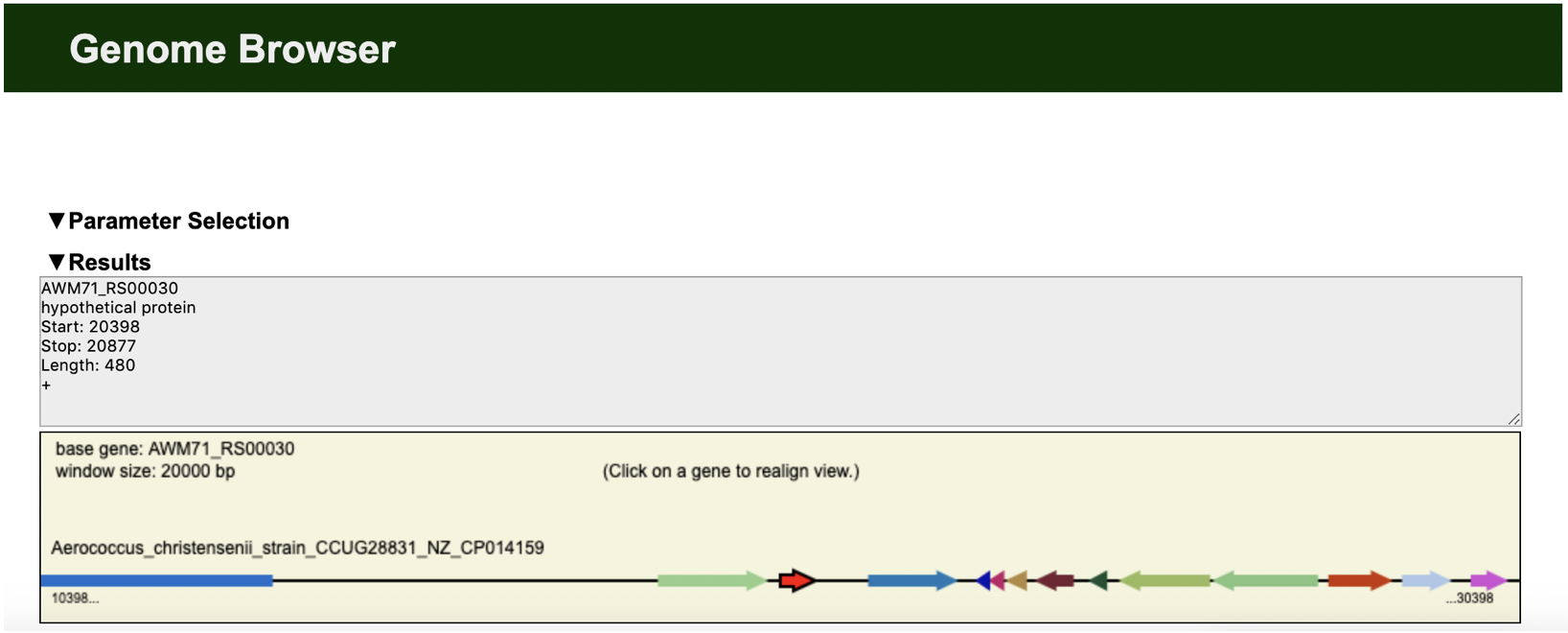 EnrichedHeatmap
EnrichedHeatmap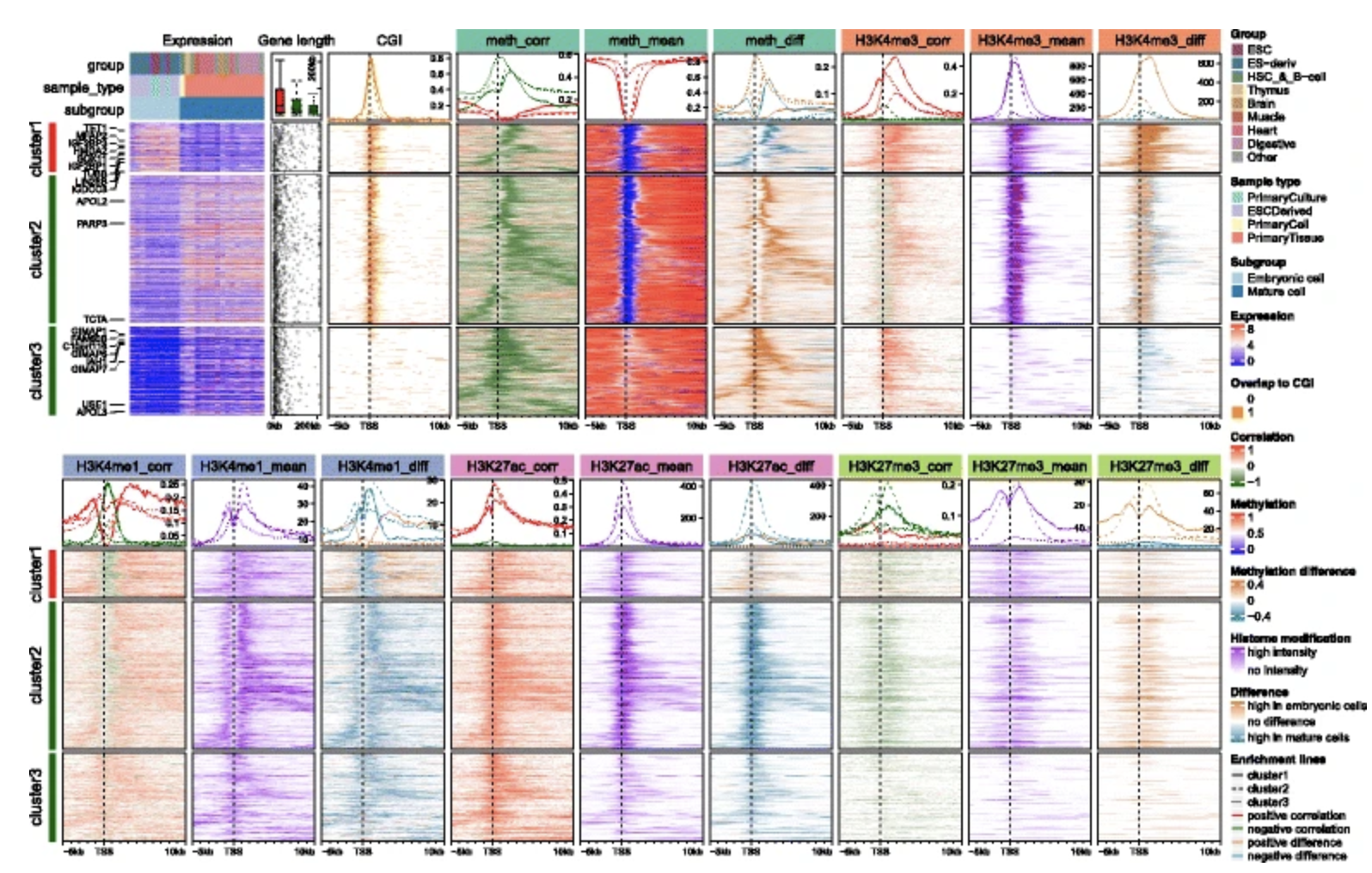 Ensembl
Ensembl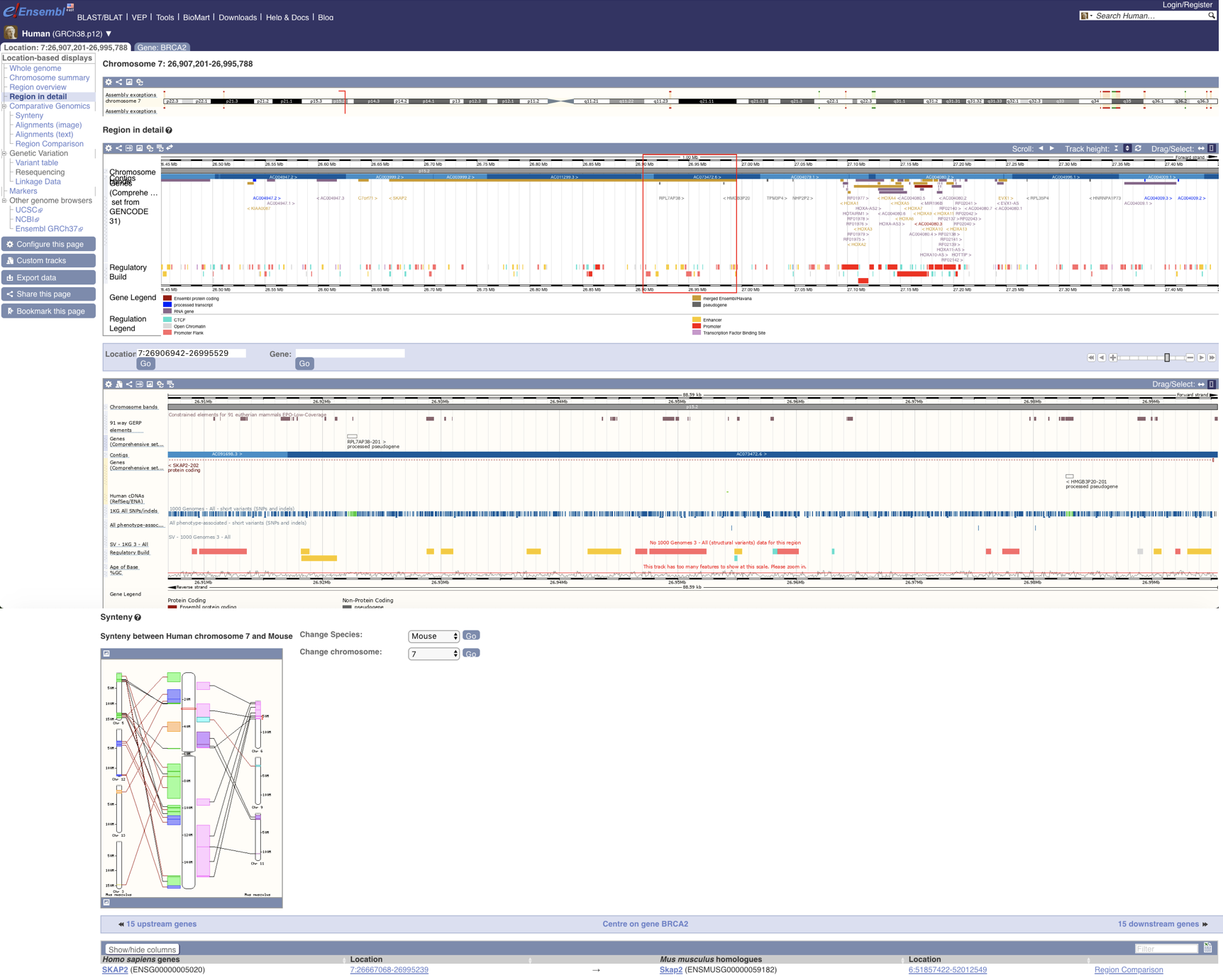 GBrowse
GBrowse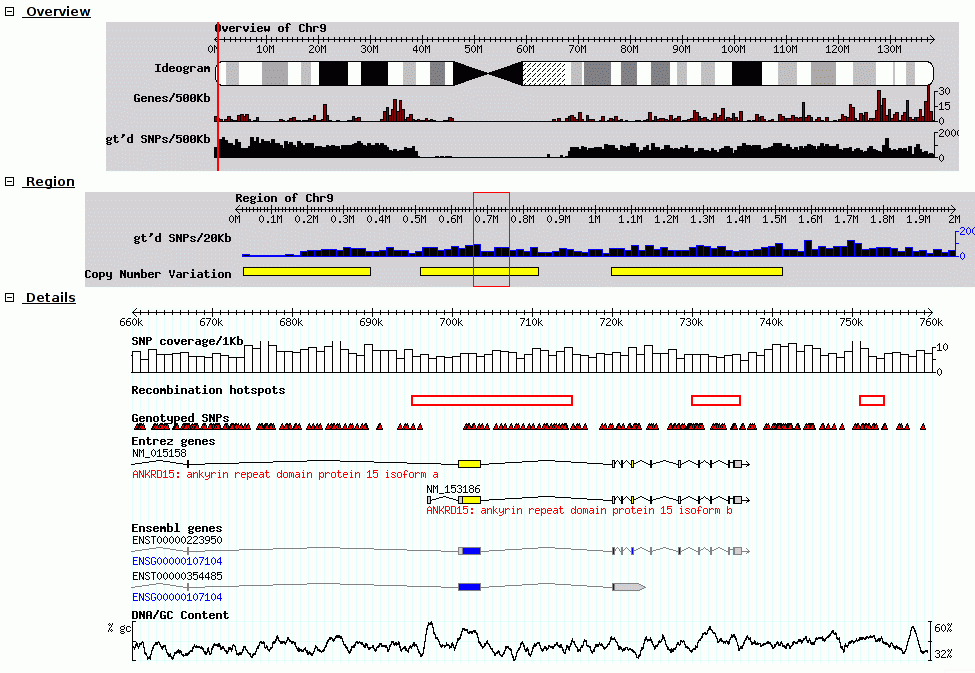 GBrowse_syn
GBrowse_syn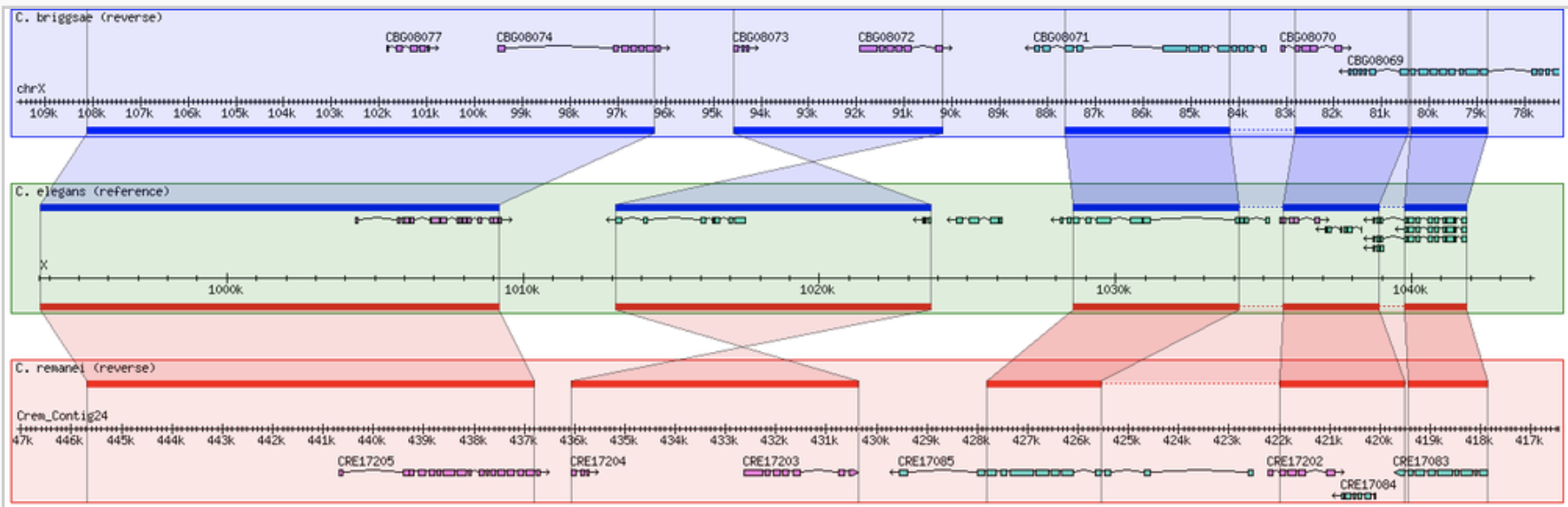 GIVE
GIVE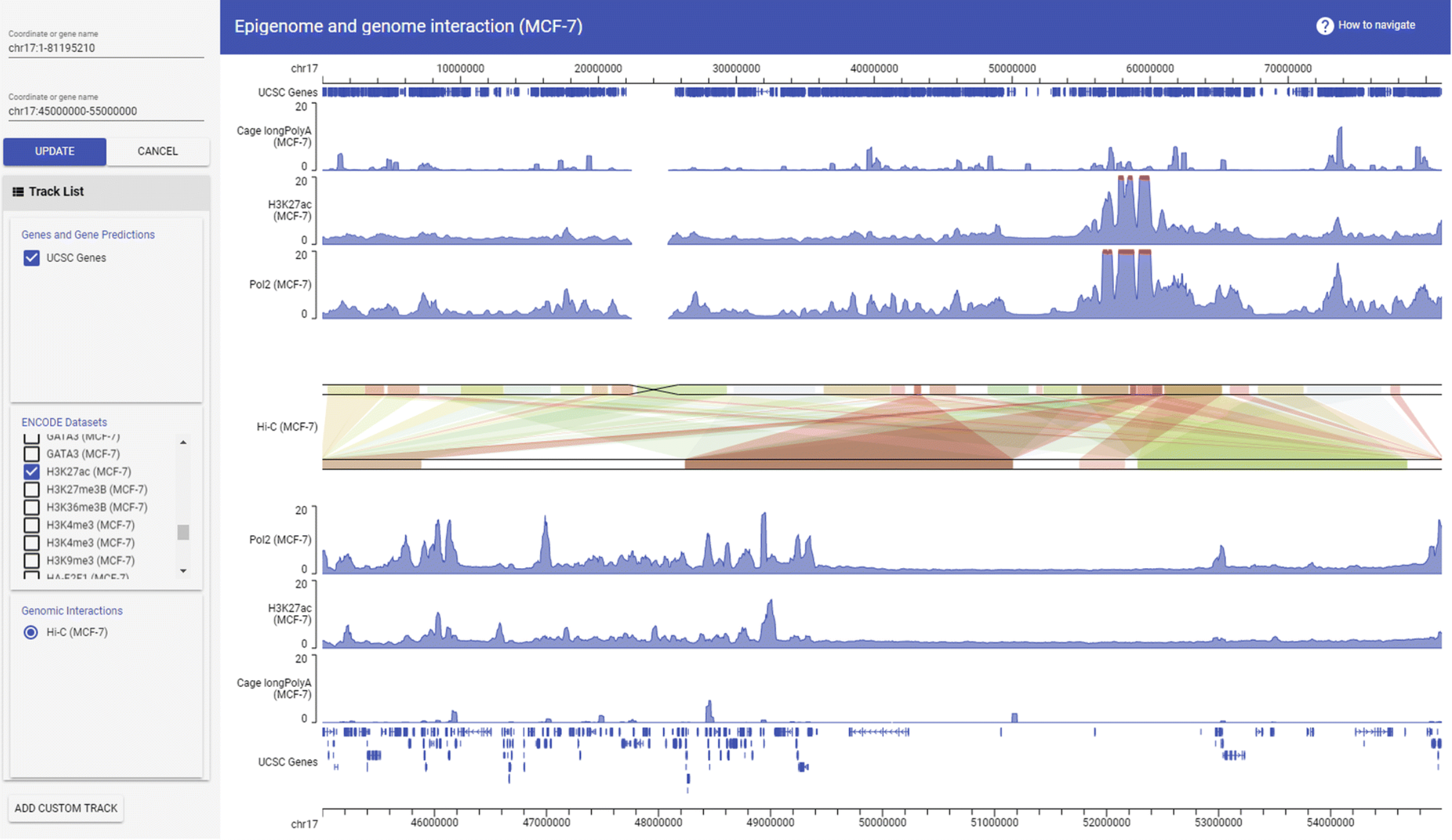 Gremlin
Gremlin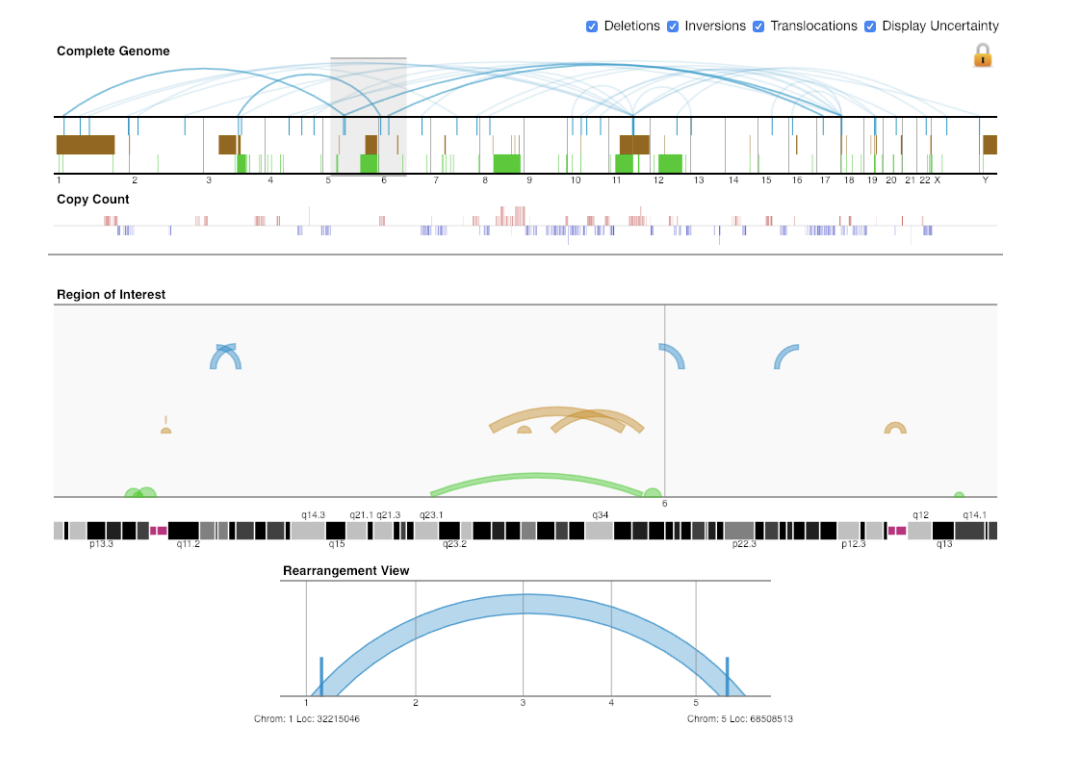 HiGlass
HiGlass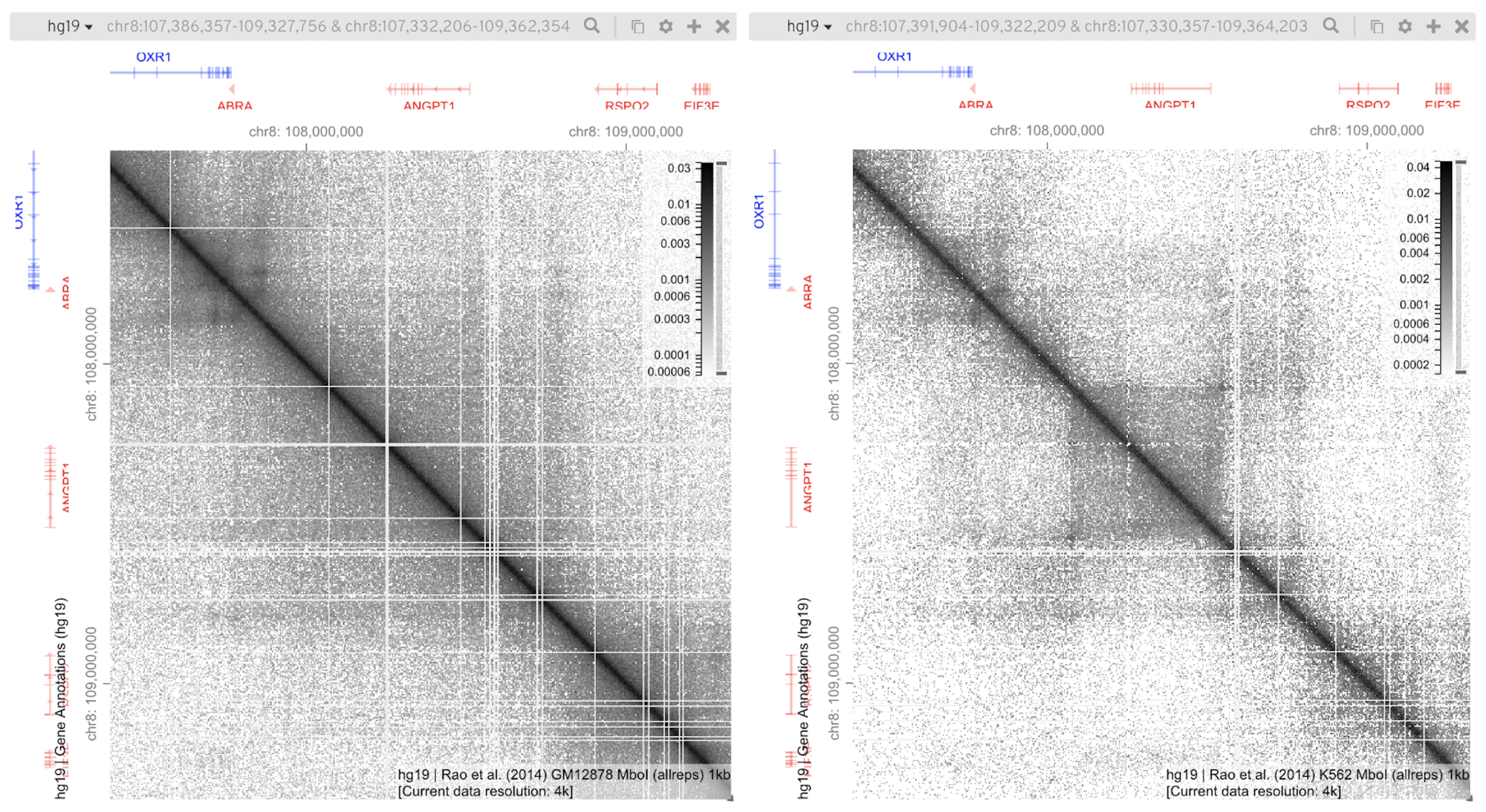 HiPiler
HiPiler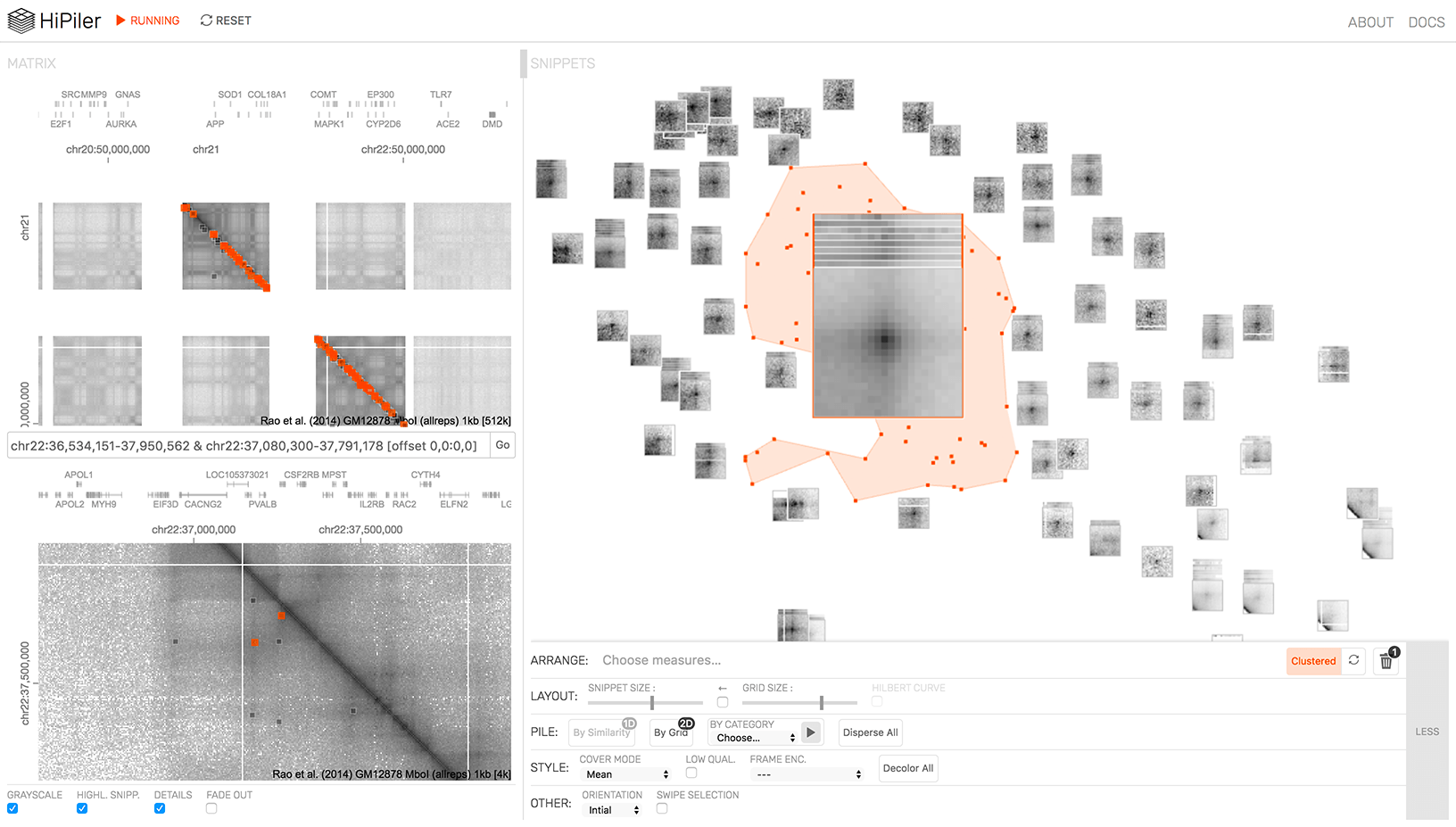 IGB
IGB Integrative Genomics Viewer
Integrative Genomics Viewer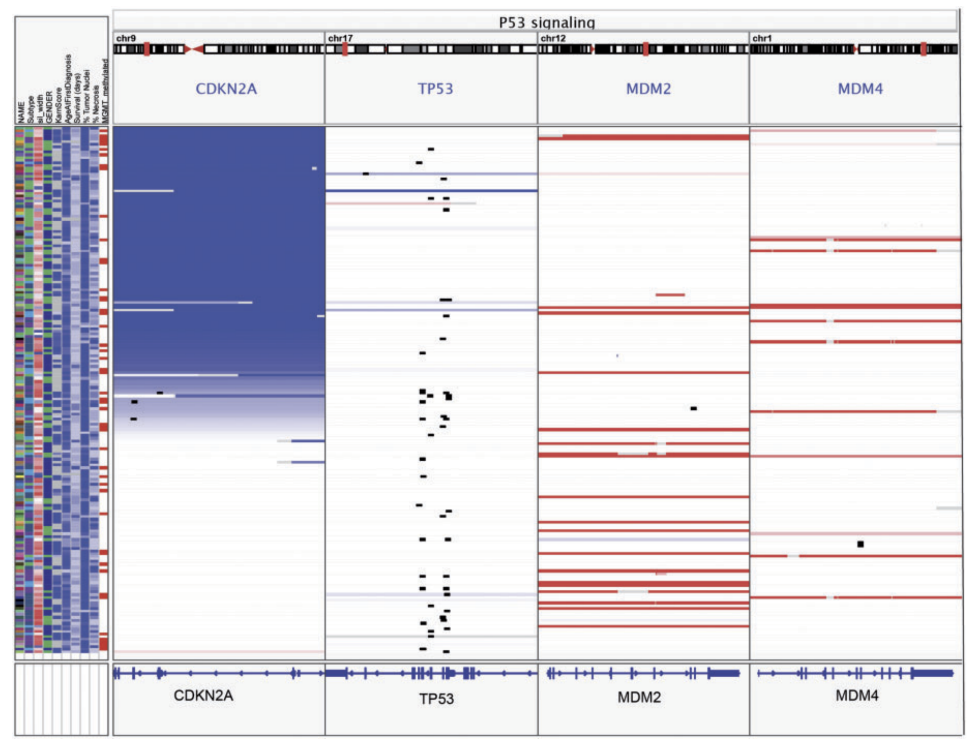 Island Viewer
Island Viewer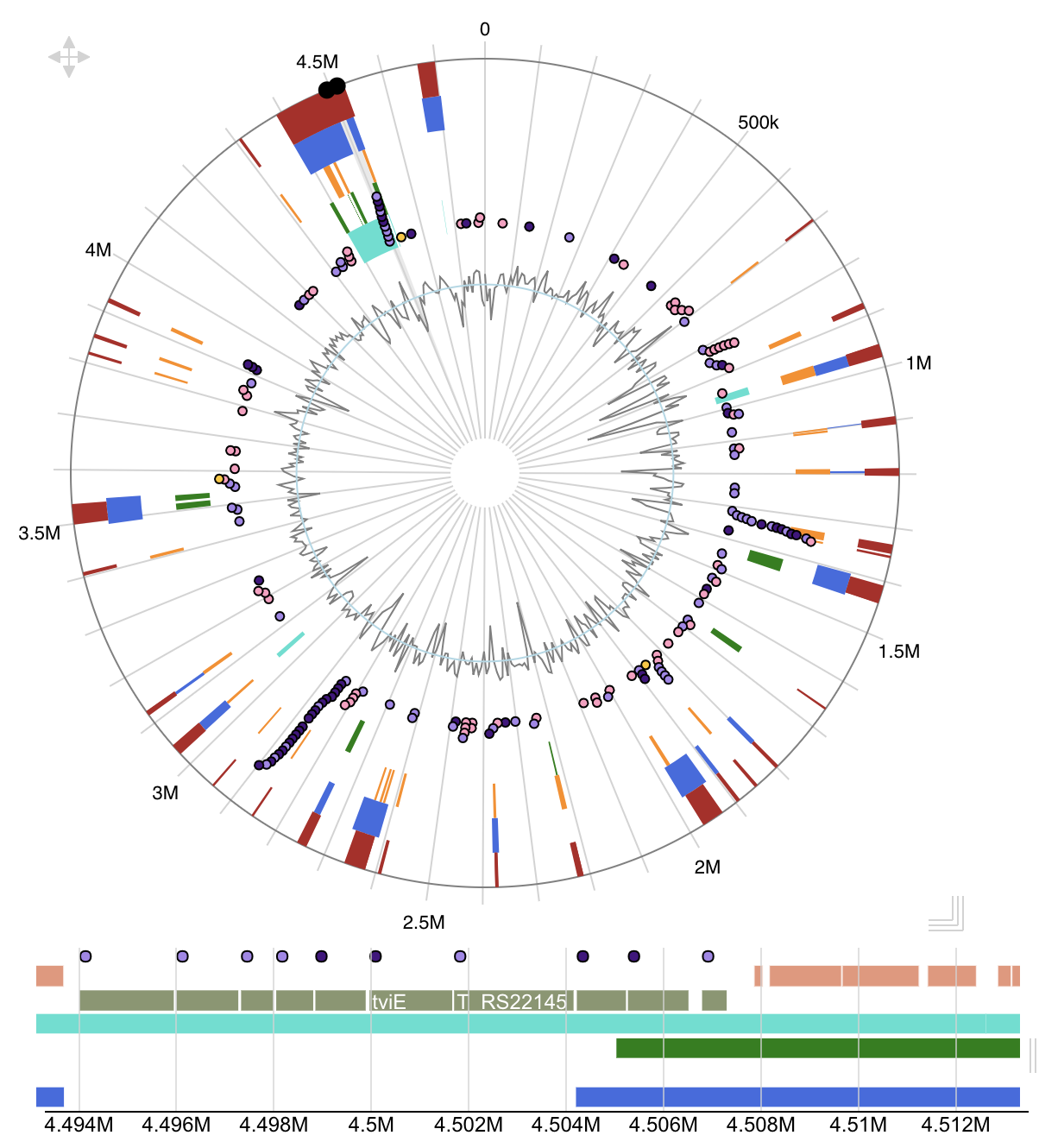 Juiceboxjs
Juiceboxjs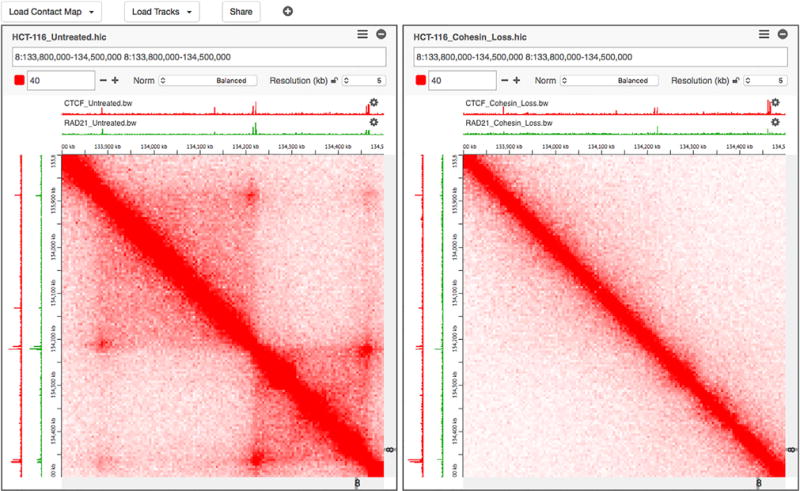 MAGI
MAGI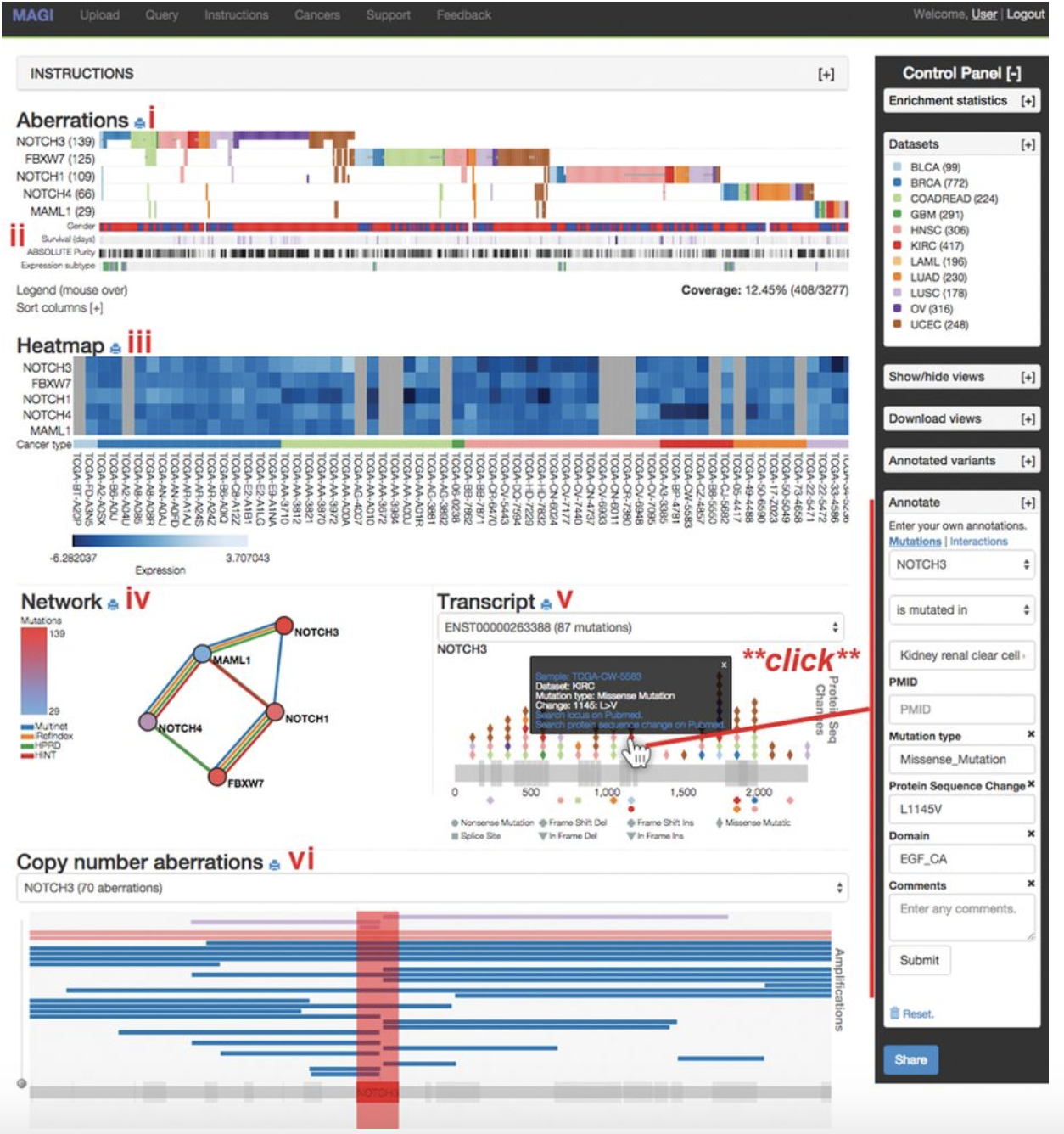 MEME
MEME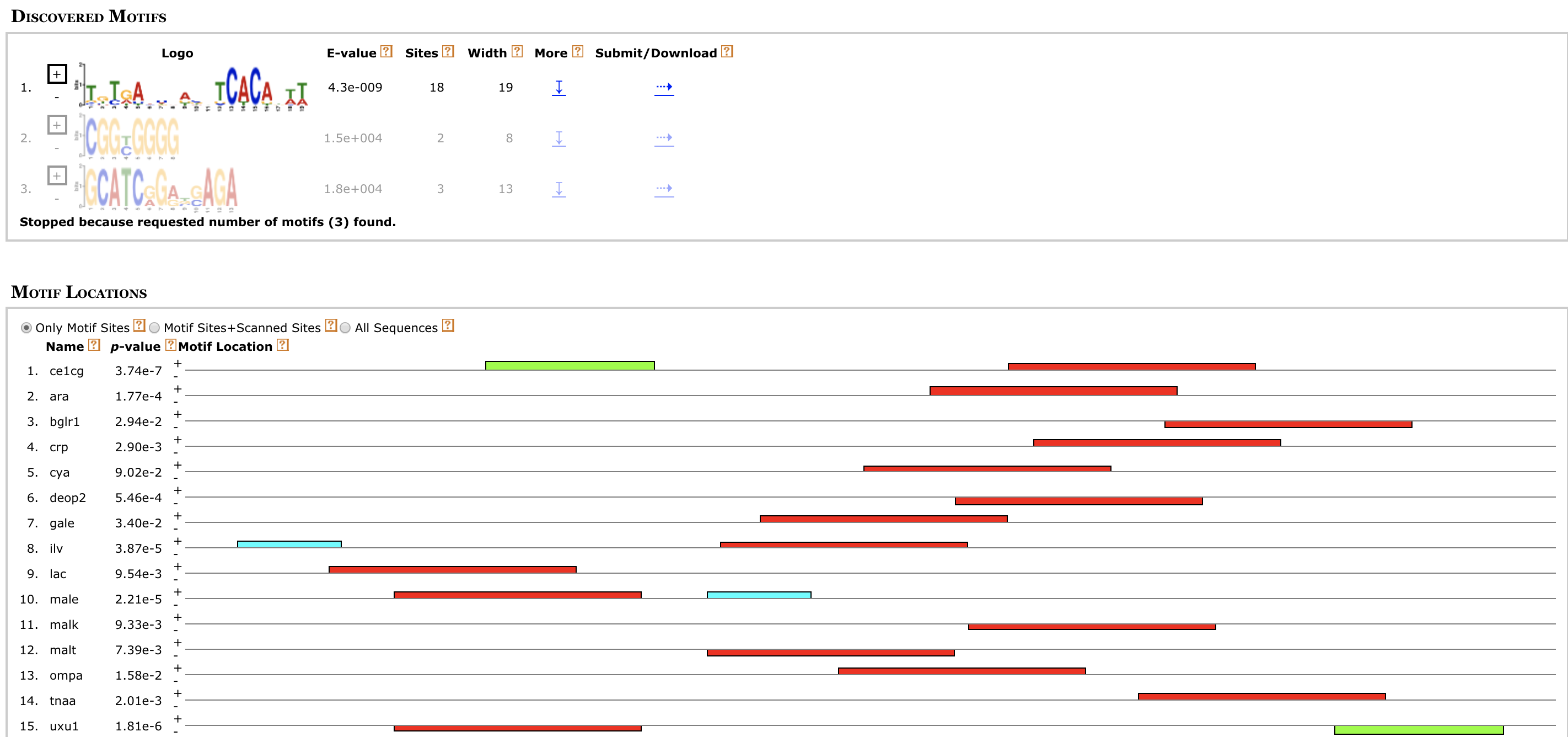 MicroScope
MicroScope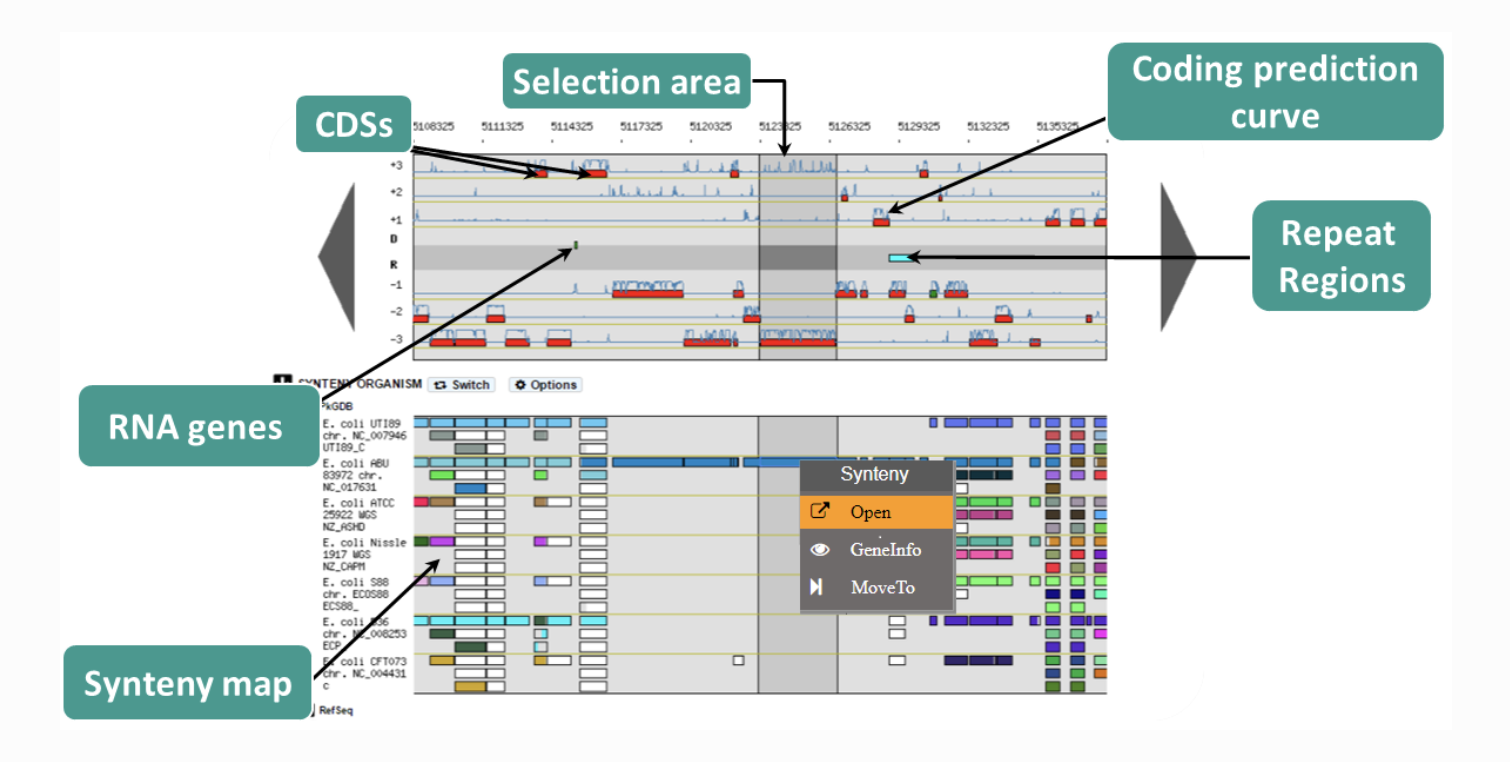 MizBee
MizBee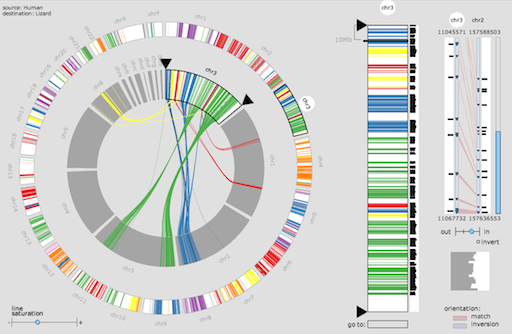 MochiView
MochiView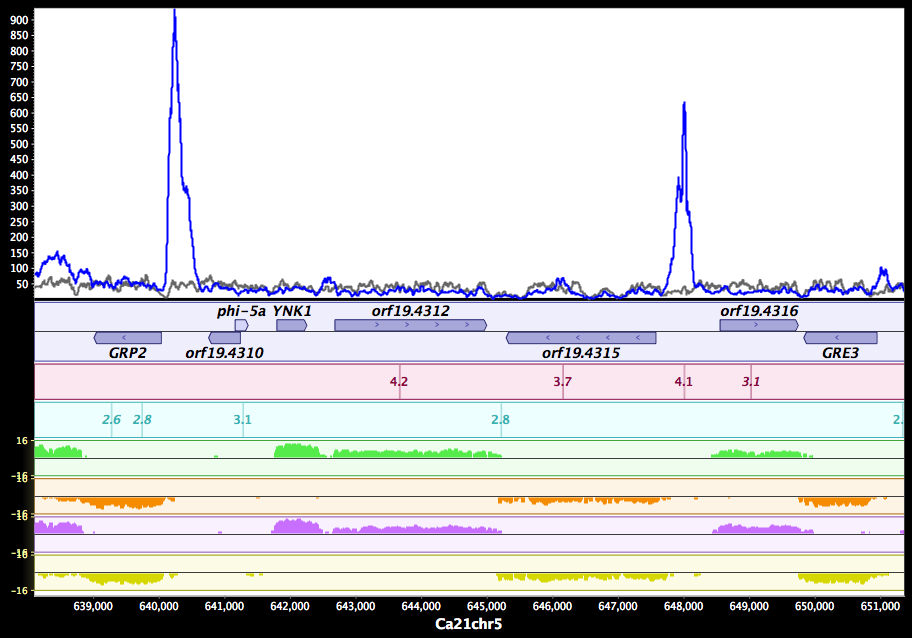 my5c
my5c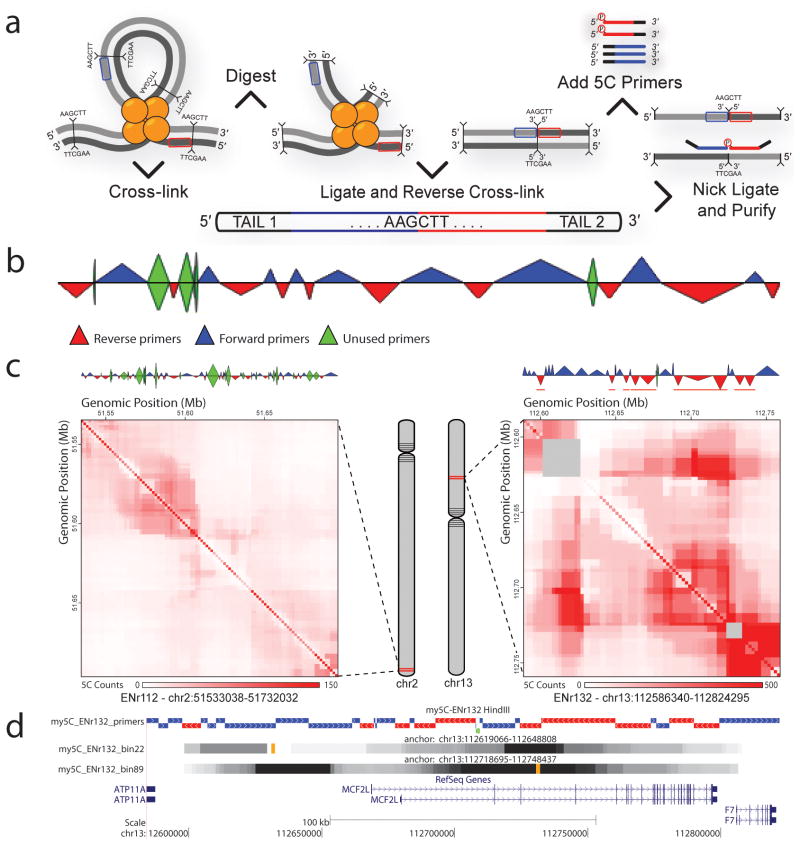 NCBI Sequence Viewer
NCBI Sequence Viewer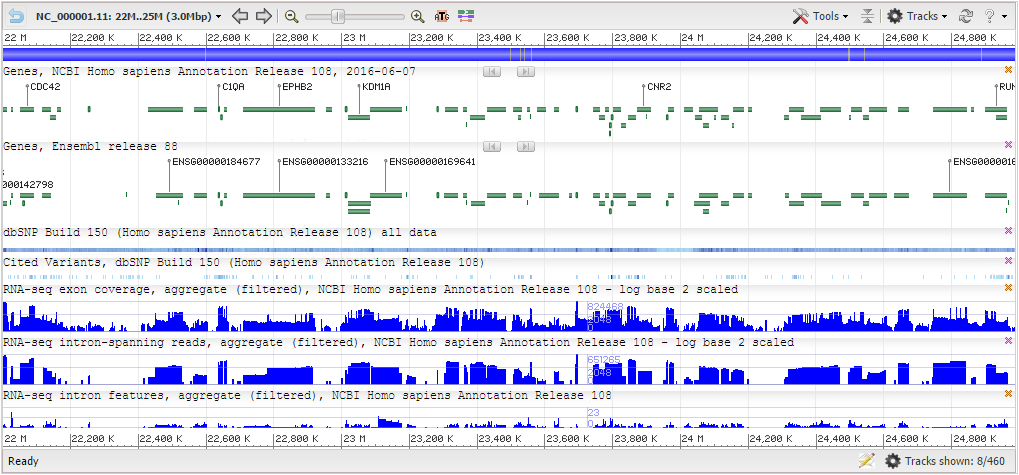 ngs.plot
ngs.plot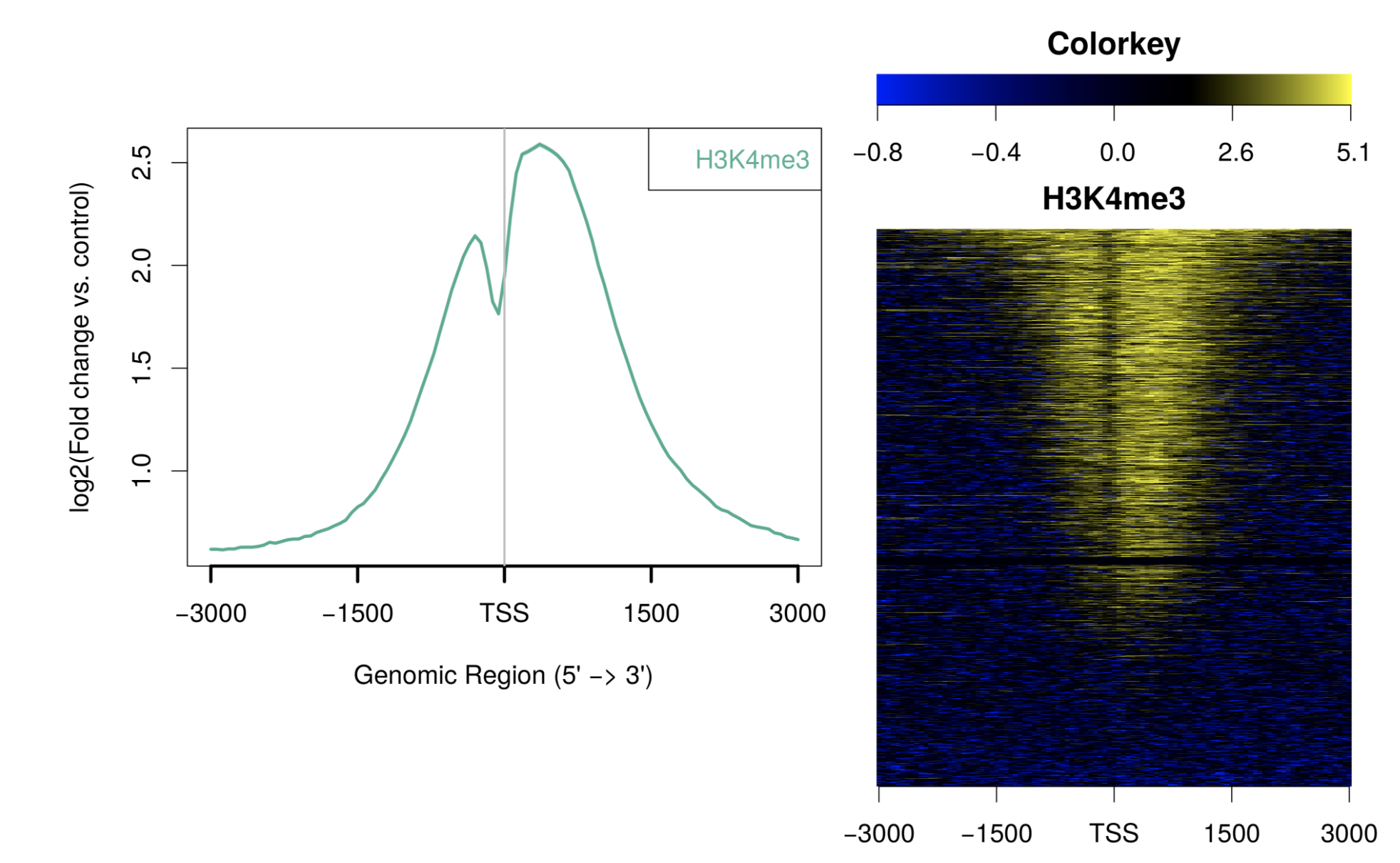 Oviz-Bio
Oviz-Bio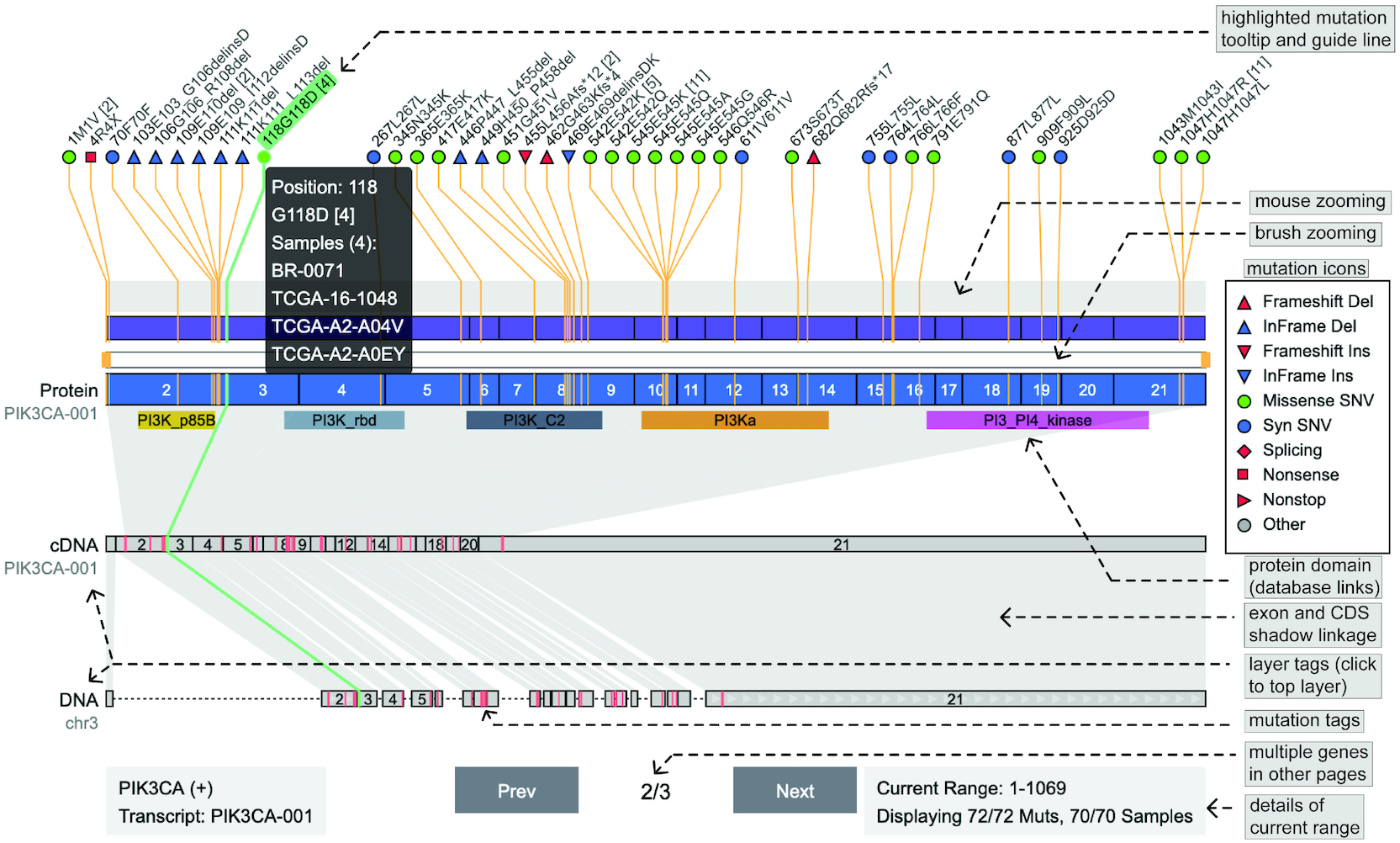 Persephone
Persephone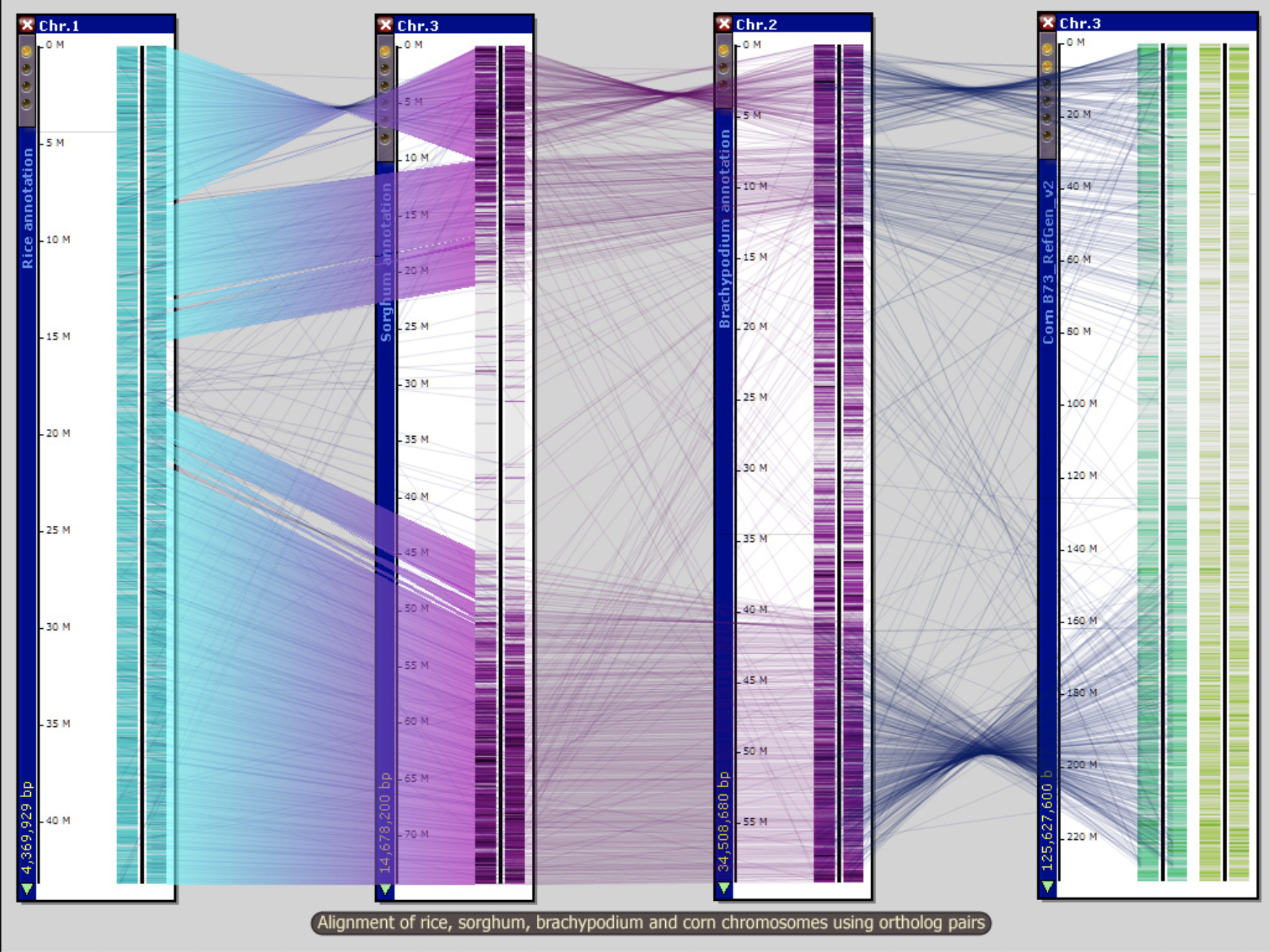 Savant Genome Browser 2
Savant Genome Browser 2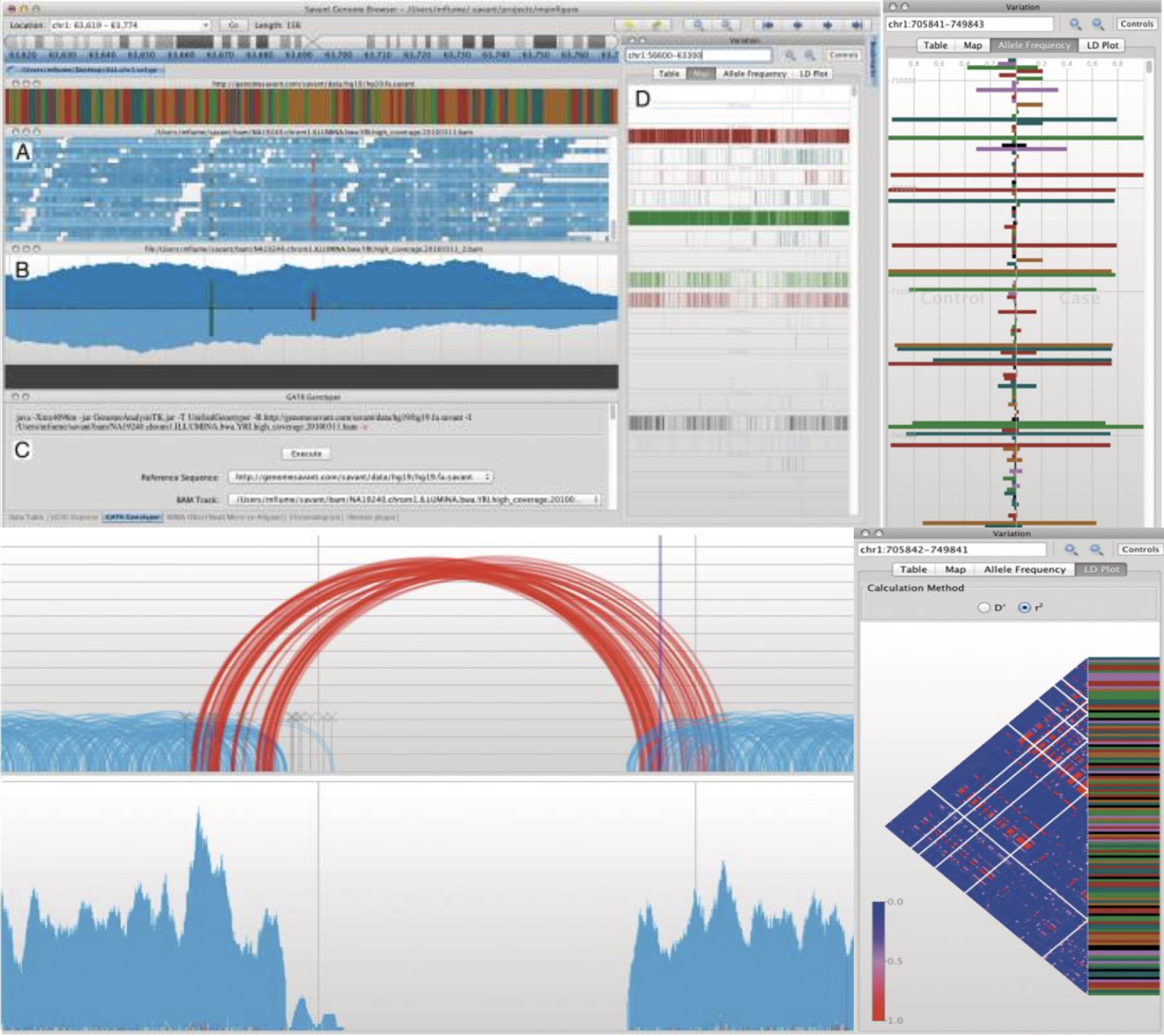 SCGV
SCGV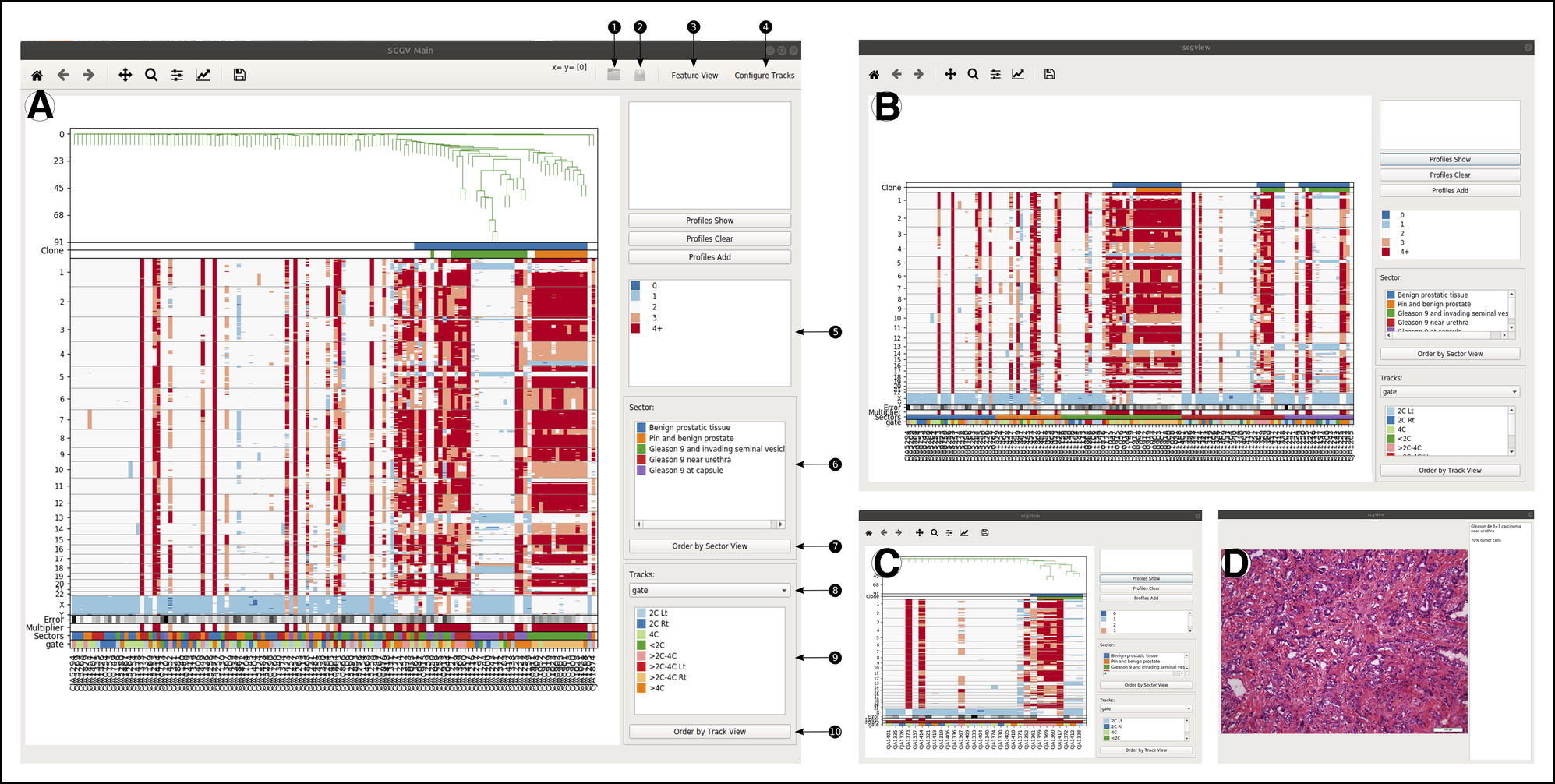 Sequence Surveyor
Sequence Surveyor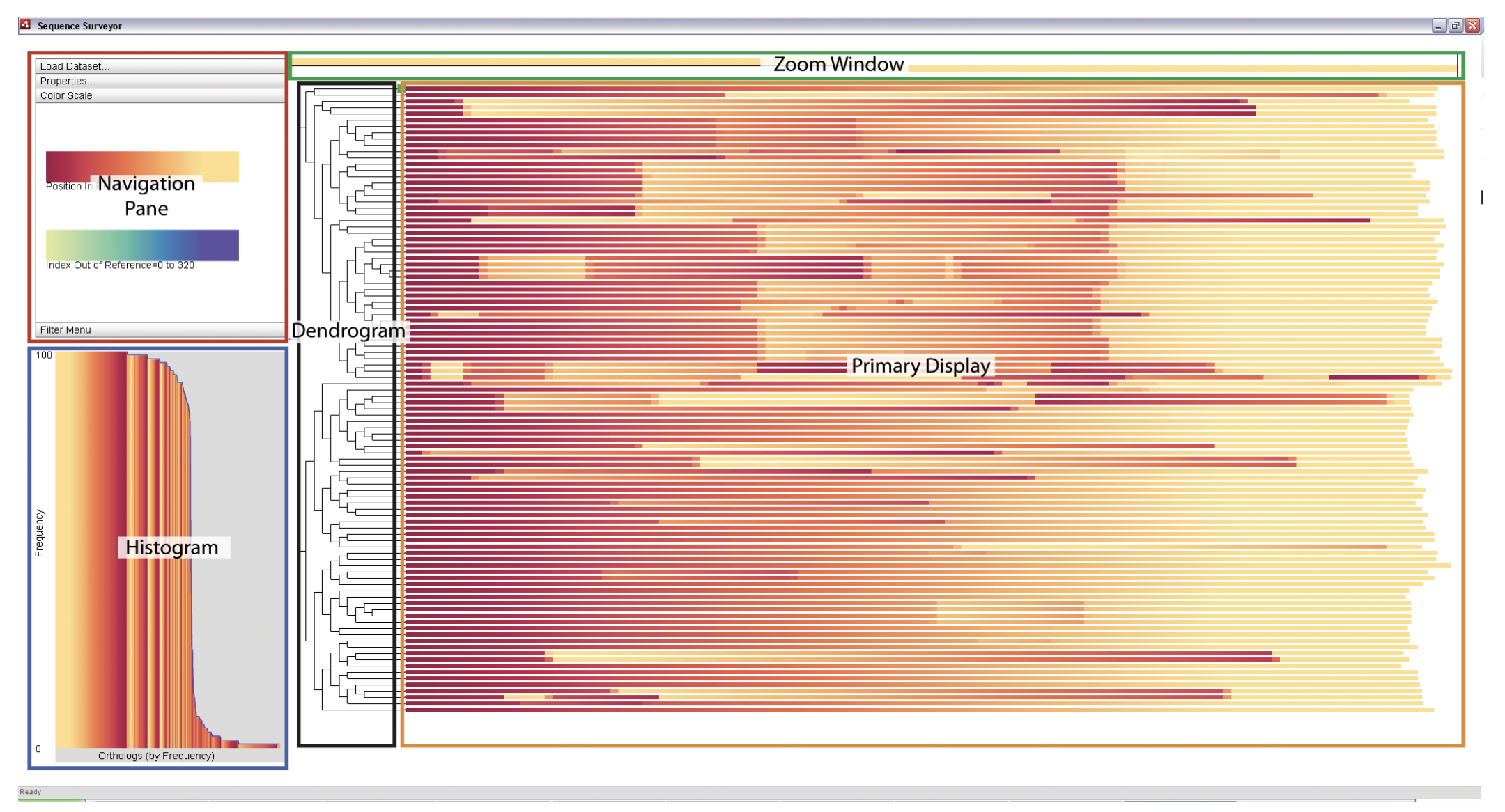 SilkDB 3.0
SilkDB 3.0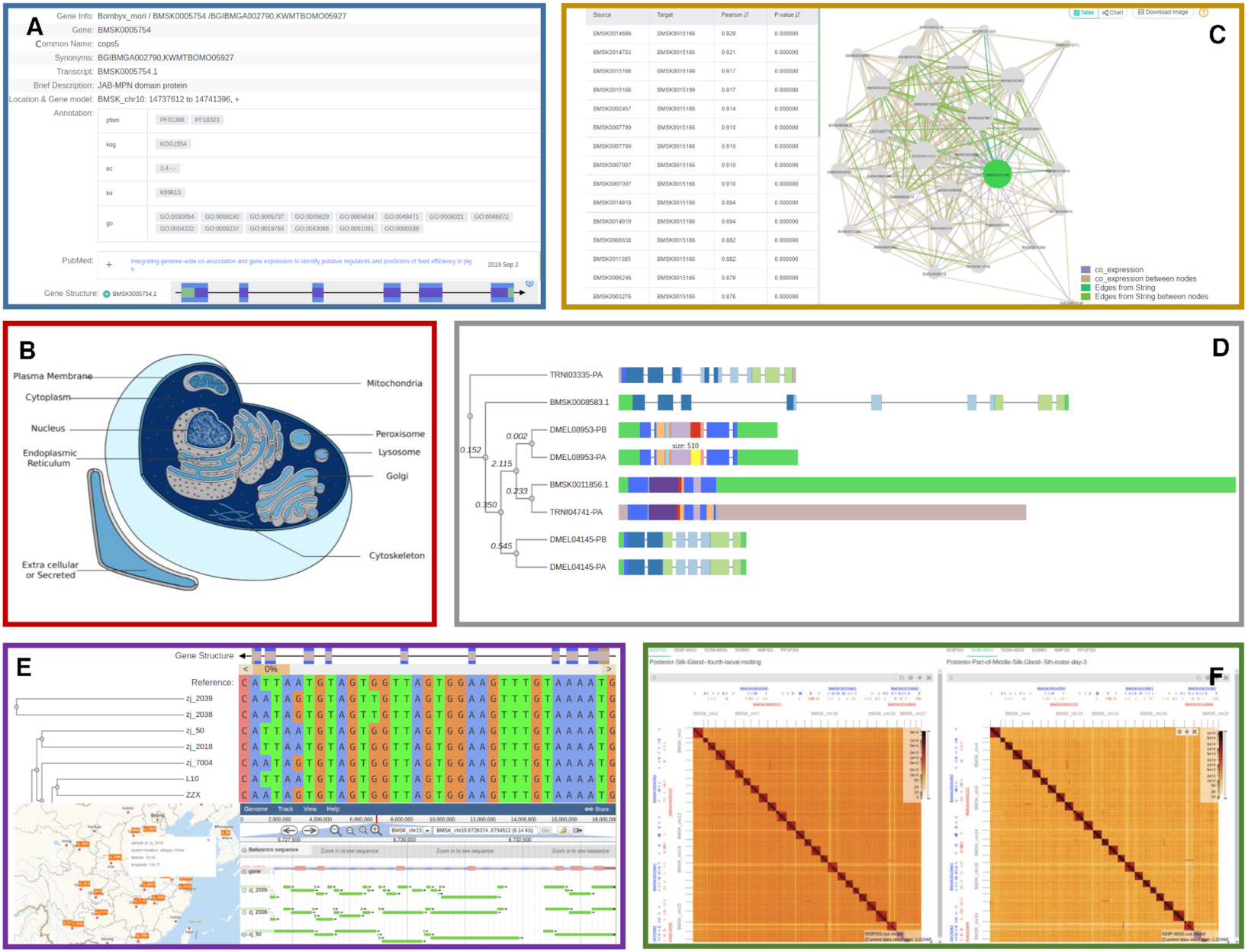 SWAV
SWAV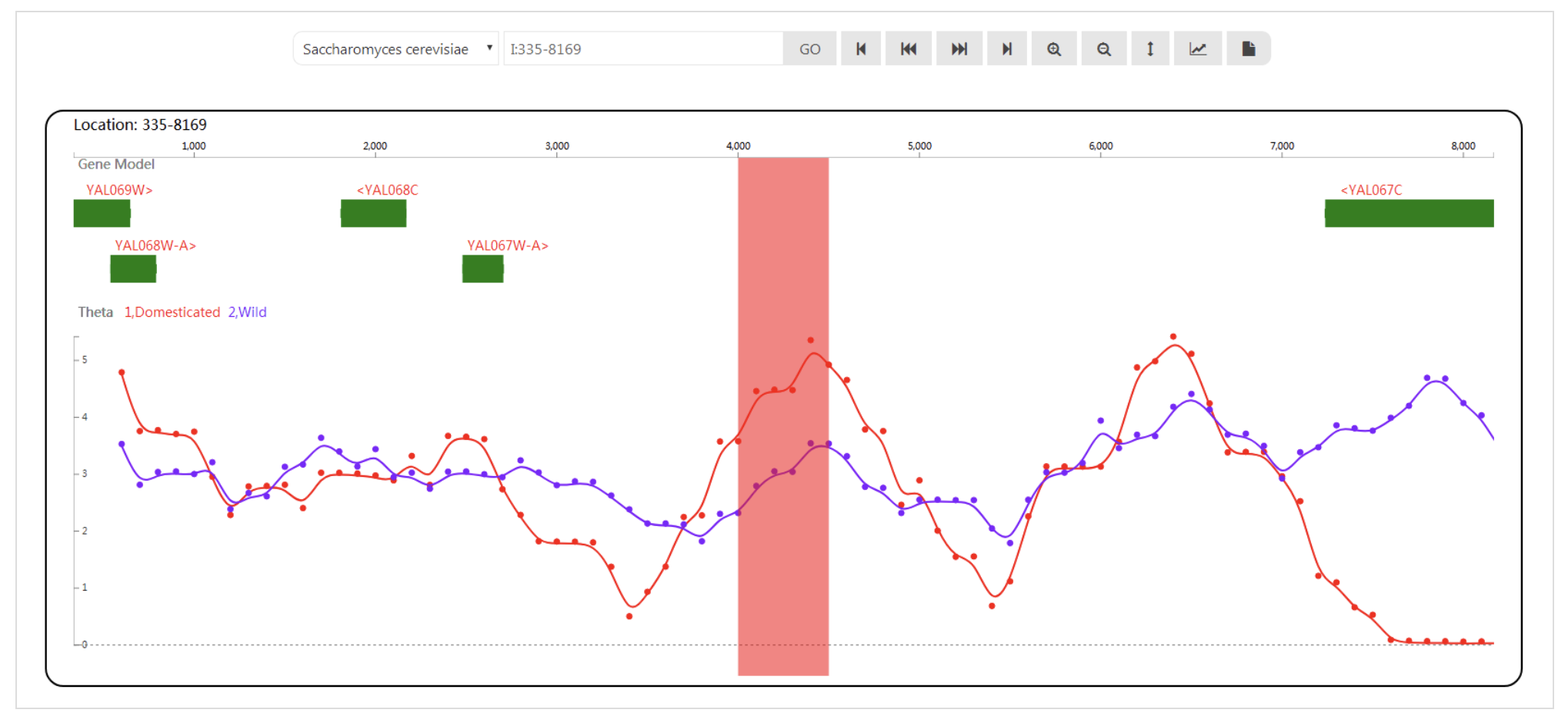 Synteny Explorer
Synteny Explorer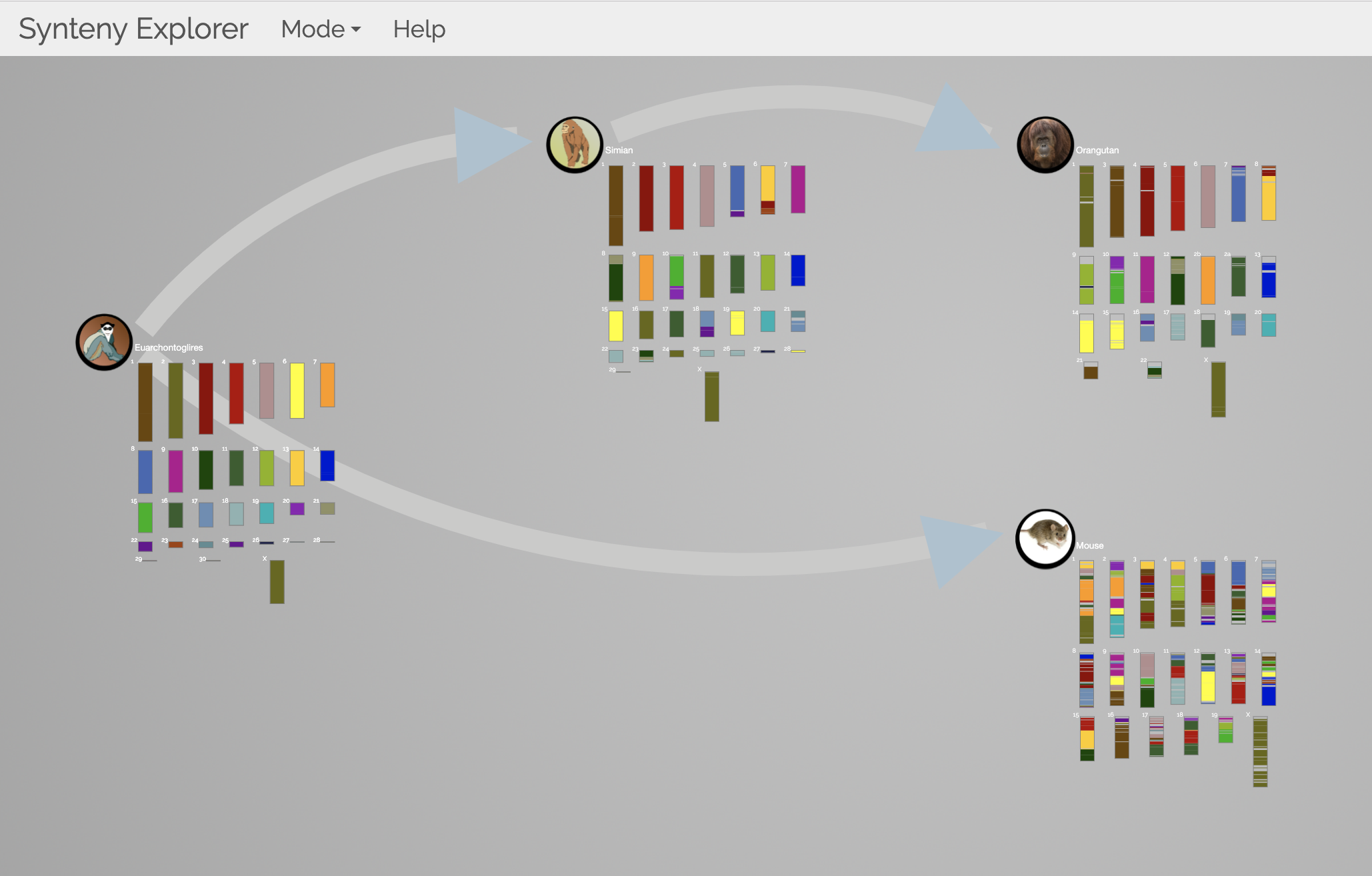 trackViewer
trackViewer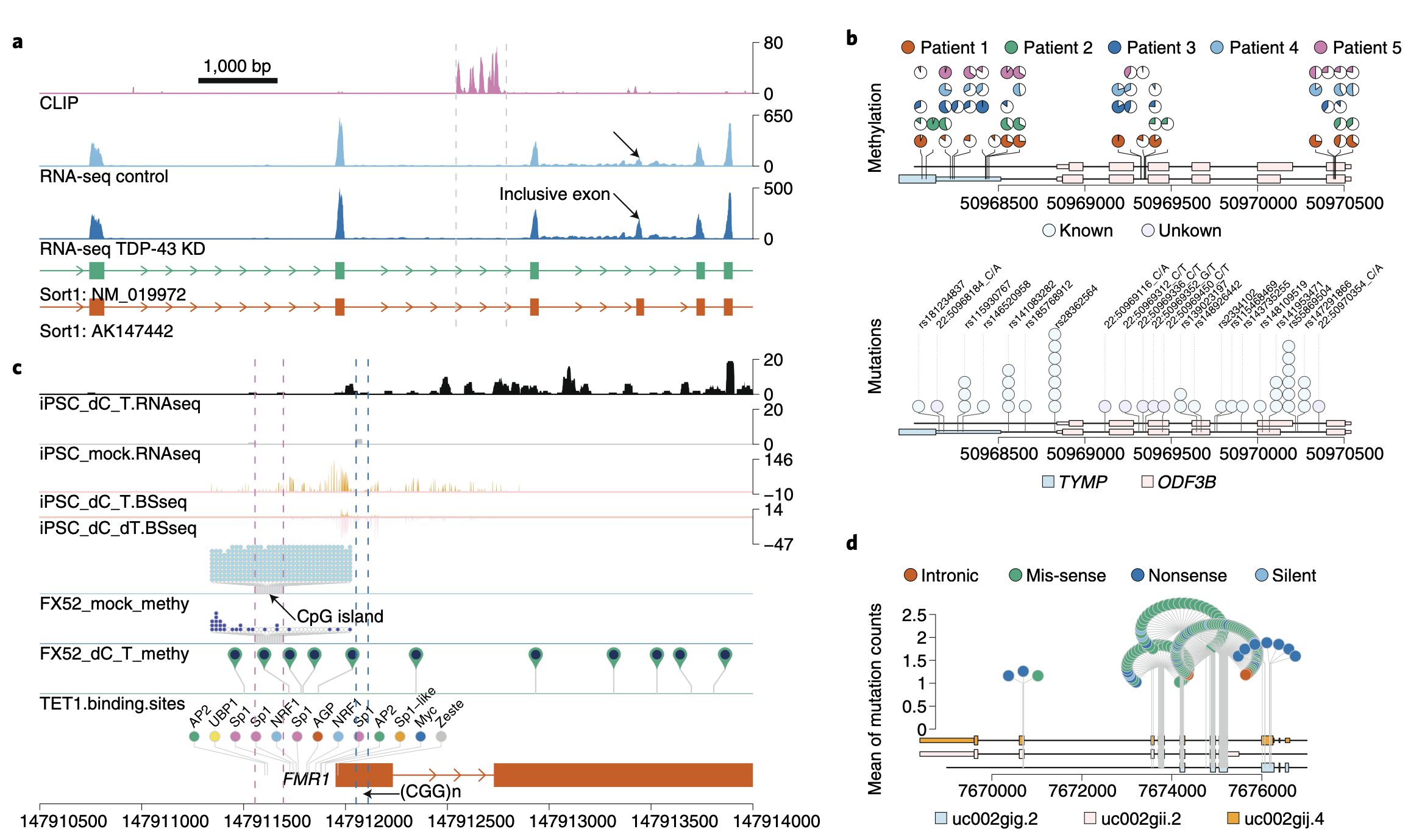 Vista Synteny
Vista Synteny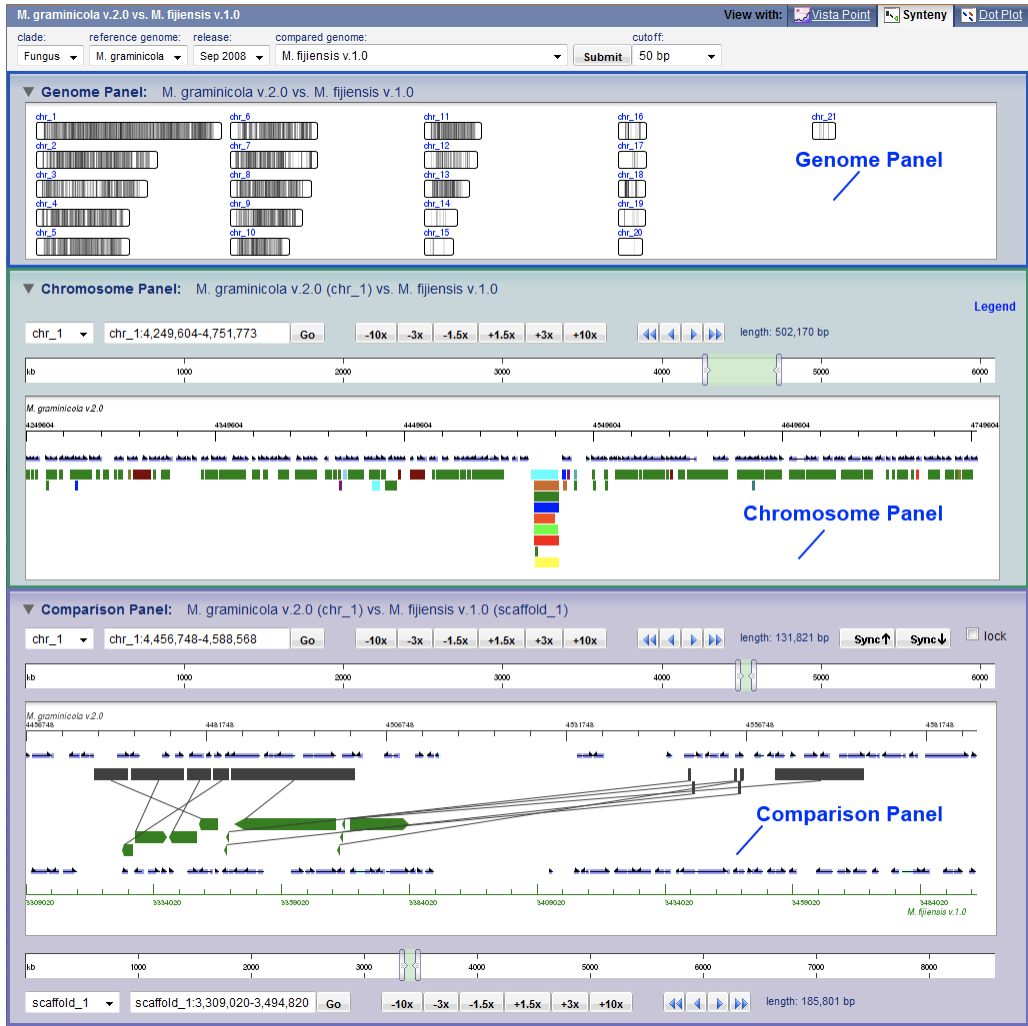 Xena
Xena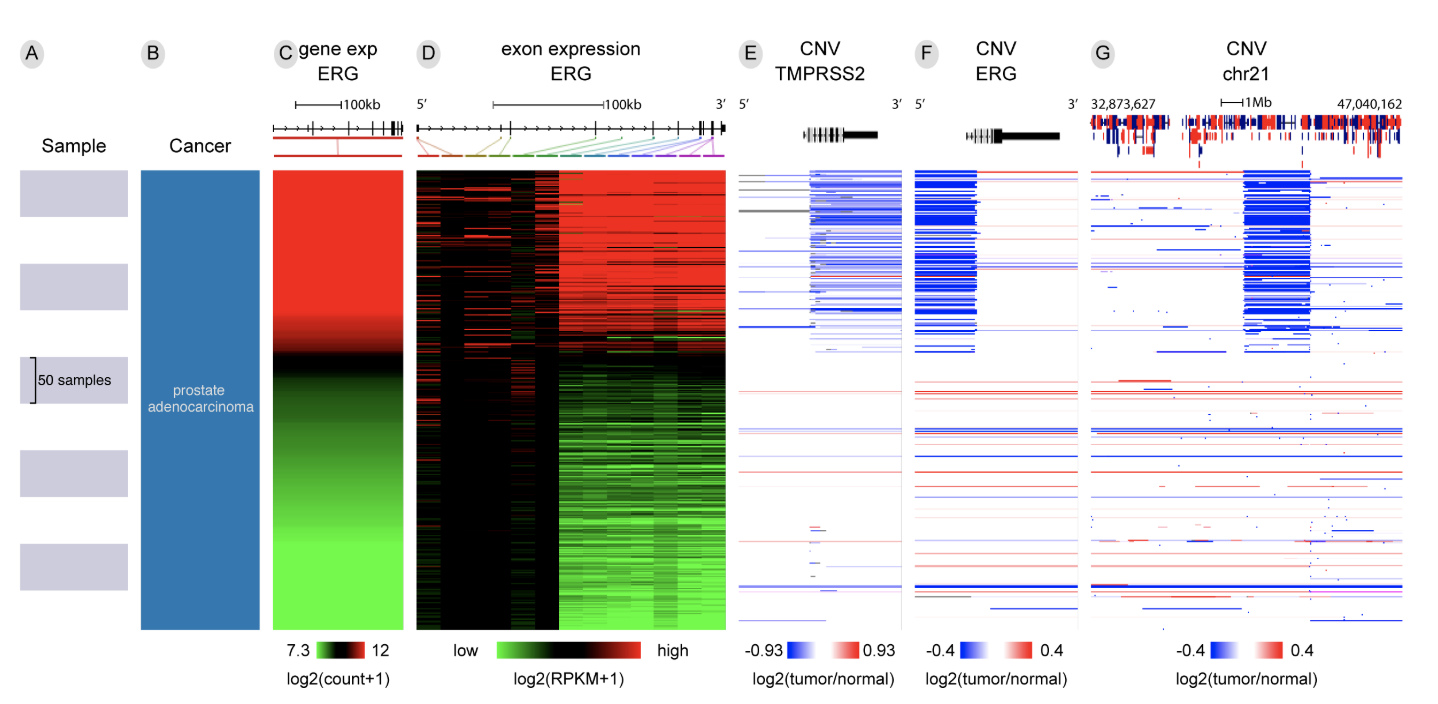
 Apollo
Apollo BedSect
BedSect CEpBrowser
CEpBrowser Cooler
Cooler CRAMER
CRAMER deepTools Heatmap
deepTools Heatmap Edgar Genome Browser
Edgar Genome Browser EnrichedHeatmap
EnrichedHeatmap Ensembl
Ensembl GBrowse
GBrowse GBrowse_syn
GBrowse_syn GIVE
GIVE Gremlin
Gremlin HiGlass
HiGlass HiPiler
HiPiler IGB
IGB Integrative Genomics Viewer
Integrative Genomics Viewer Island Viewer
Island Viewer Juiceboxjs
Juiceboxjs MAGI
MAGI MEME
MEME MicroScope
MicroScope MizBee
MizBee MochiView
MochiView my5c
my5c NCBI Sequence Viewer
NCBI Sequence Viewer ngs.plot
ngs.plot Oviz-Bio
Oviz-Bio Persephone
Persephone Savant Genome Browser 2
Savant Genome Browser 2 SCGV
SCGV Sequence Surveyor
Sequence Surveyor SilkDB 3.0
SilkDB 3.0 SWAV
SWAV Synteny Explorer
Synteny Explorer trackViewer
trackViewer Vista Synteny
Vista Synteny Xena
Xena