GBrowse
 http://gmod.org/wiki/GBrowse#Screenshots
http://gmod.org/wiki/GBrowse#Screenshots
Tool
Paper
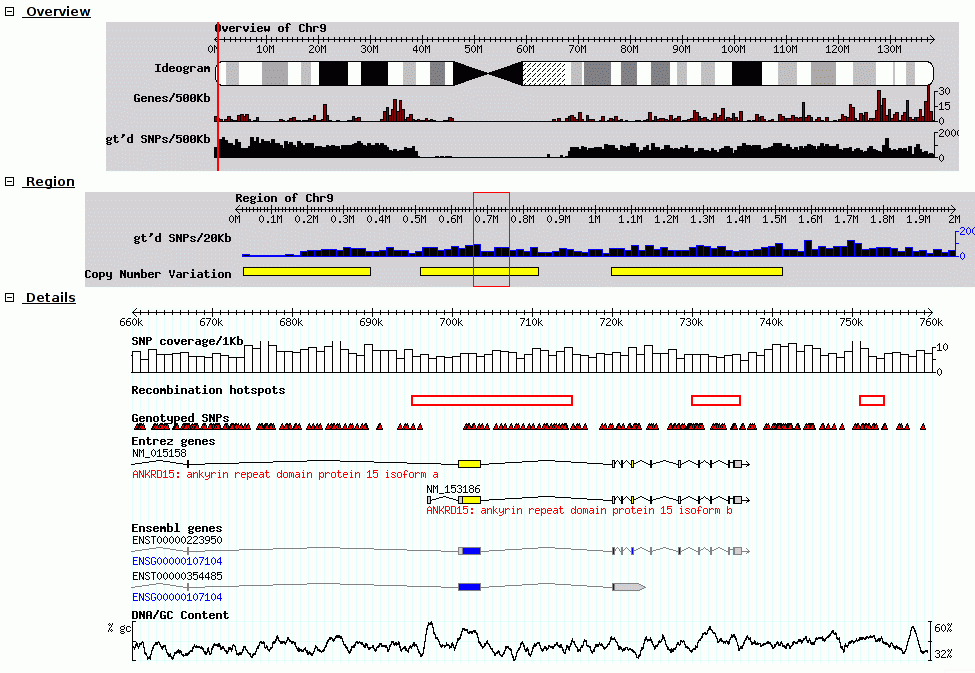
Using the Generic Genome Browser (GBrowse).
Donlin MJ. Using the Generic Genome Browser (GBrowse). Curr Protoc Bioinformatics. 2009;Chapter 9: Unit 9.9.
Cited by: 127
Abstract
A genome browser is software that allows users to visualize DNA, protein, or other sequence features within the context of a reference sequence, such as a chromosome or contig. The Generic Genome Browser (GBrowse) is an open‐source browser developed as part of the Generic Model Organism Database project (Stein et al., 2002). GBrowse can be configured to display genomic sequence features for any organism and is the browser used for the model organisms Drosophila melanogaster (Grumbling and Strelets, 2006) and Caenorhabditis elegans (Schwarz et al., 2006), among others. The software package can be downloaded from the Web and run on a Windows, Mac OS X, or Unix‐type system. Version 1.64, as described in the original protocol, was released in November 2005, but the software is under active development and new versions are released about every six months. This update includes instructions on updating existing data sources with new files from NCBI. Curr. Protoc. Bioinform. 28:9.9.1‐9.9.25. © 2009 by John Wiley & Sons, Inc
 http://gmod.org/wiki/GBrowse#Screenshots
http://gmod.org/wiki/GBrowse#Screenshots