SpliceGrapher
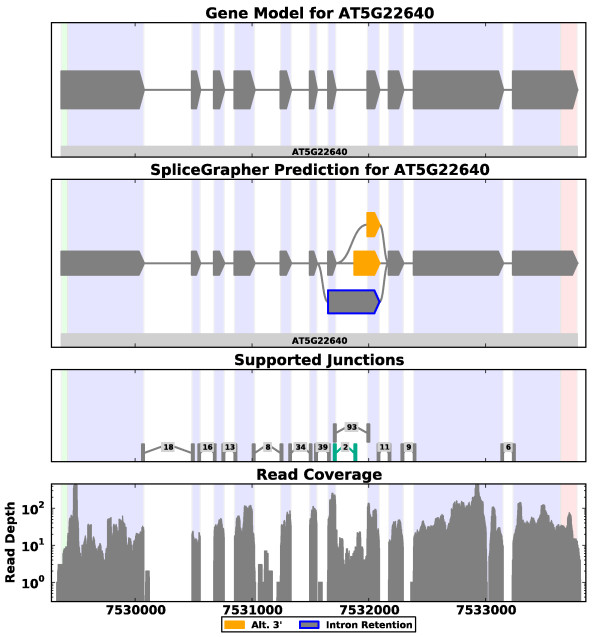 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3334585/figure/F1/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3334585/figure/F1/
Methods
linear single view single scale single focus segregated partial abstraction no arrangement no interconnection segment sparse typeTool
Tool:
SpliceGrapherTool Link:
http://splicegrapher.sourceforge.net/Documentation:
http://splicegrapher.sourceforge.net/userguide.htmlPaper
SpliceGrapher: detecting patterns of alternative splicing from RNA-Seq data in the context of gene models and EST data.
Rogers MF, Thomas J, Reddy AS, Ben-Hur A. SpliceGrapher: detecting patterns of alternative splicing from RNA-Seq data in the context of gene models and EST data. Genome Biol. genomebiology.biomedcentral.com; 2012;13: R4.
Cited by: 103