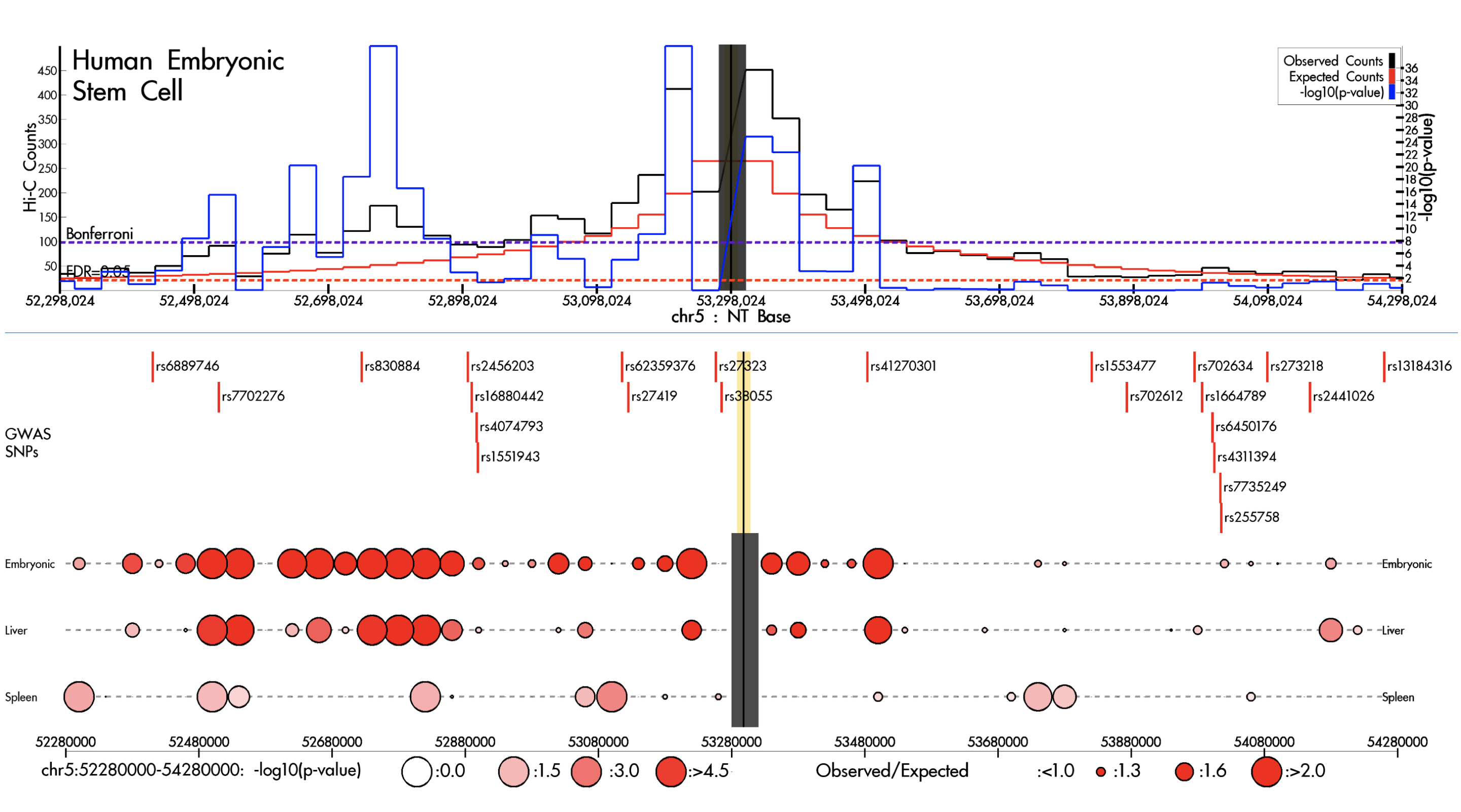
HUGIn
 https://yunliweb.its.unc.edu/HUGIn/#plots
https://yunliweb.its.unc.edu/HUGIn/#plots
Methods
linear single view single scale single focus segregated no abstraction no arrangement between interconnection segment sparse type segment contiguous type point sparse type point contiguous typeTool
Tool:
HUGInTool Link:
http://yunliweb.its.unc.edu/HUGInDocumentation:
https://yunliweb.its.unc.edu/HUGIn/tutorial_page.phpPaper
HUGIn: Hi-C Unifying Genomic Interrogator.
Martin JS, Xu Z, Reiner AP, Mohlke KL, Sullivan P, Ren B, et al. HUGIn: Hi-C Unifying Genomic Interrogator. Bioinformatics. academic.oup.com; 2017;33: 3793–3795.
Cited by: 31