Variant View
 http://www.cs.ubc.ca/labs/imager/tr/2013/VariantView/VariantViewSoftware/variant_view/VariantView.html
http://www.cs.ubc.ca/labs/imager/tr/2013/VariantView/VariantViewSoftware/variant_view/VariantView.html
Methods
linear single view single scale single focus segregated no abstraction no arrangement no interconnection segment sparse type point sparse typeTool
| Access Format | standalone app |
| Supported Files | other |
| License | New BSD license |
| Tool name | Variant View |
| Tool Link | https://www.cs.ubc.ca/labs/imager/tr/2013/VariantView/VariantViewSoftware/ |
| Documentation | https://www.cs.ubc.ca/labs/imager/tr/2013/VariantView/VariantViewSoftware/variant_view/READ_ME.txt |
Paper
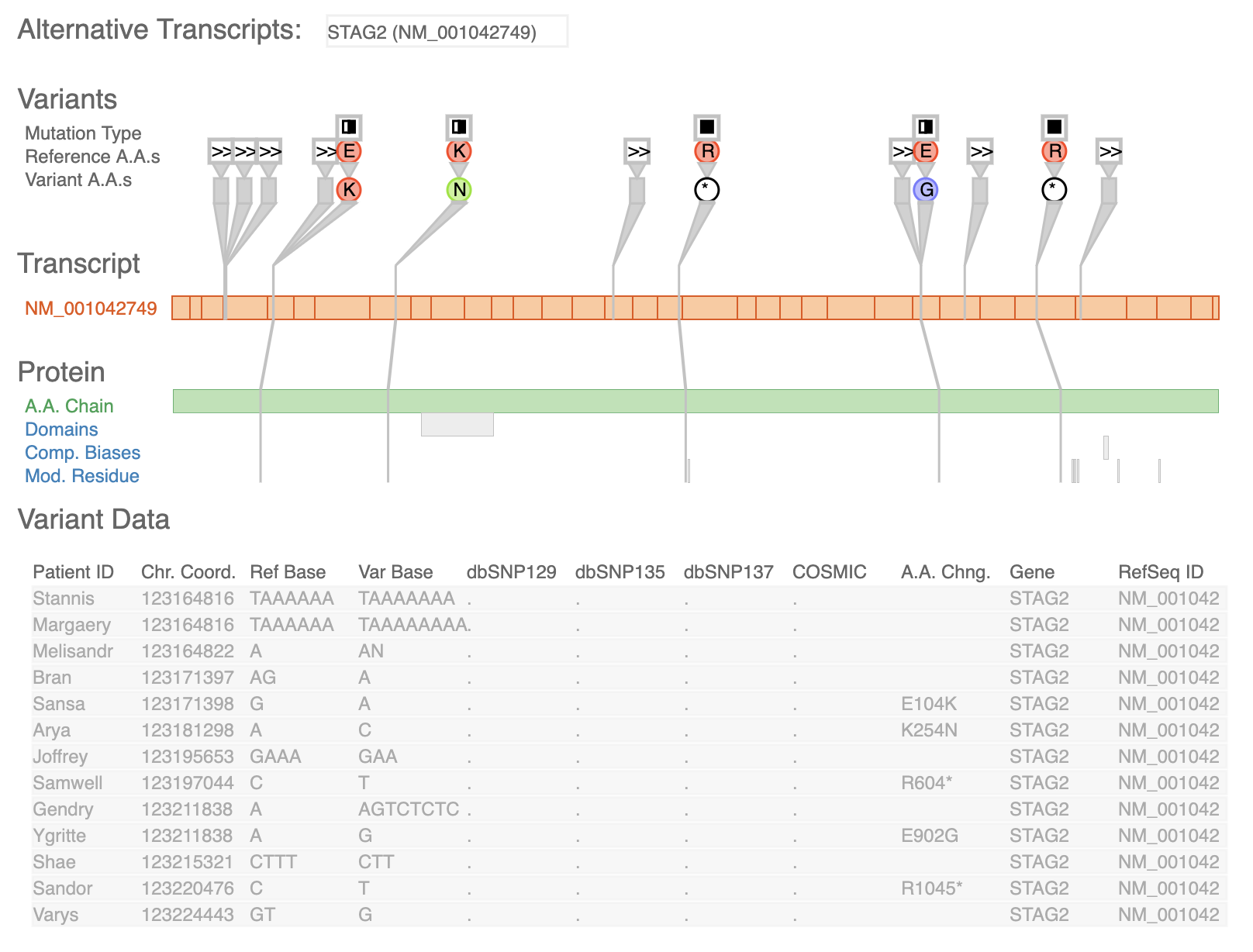
Variant view: visualizing sequence variants in their gene context.
Ferstay JA, Nielsen CB, Munzner T. Variant view: visualizing sequence variants in their gene context. IEEE Trans Vis Comput Graph. 2013;19: 2546–2555.
Abstract
Scientists use DNA sequence differences between an individual's genome and a standard reference genome to study the genetic basis of disease. Such differences are called sequence variants, and determining their impact in the cell is difficult because it requires reasoning about both the type and location of the variant across several levels of biological context. In this design study, we worked with four analysts to design a visualization tool supporting variant impact assessment for three different tasks. We contribute data and task abstractions for the problem of variant impact assessment, and the carefully justified design and implementation of the Variant View tool. Variant View features an information-dense visual encoding that provides maximal information at the overview level, in contrast to the extensive navigation required by currently-prevalent genome browsers. We provide initial evidence that the tool simplified and accelerated workflows for these three tasks through three case studies. Finally, we reflect on the lessons learned in creating and refining data and task abstractions that allow for concise overviews of sprawling information spaces that can reduce or remove the need for the memory-intensive use of navigation.