Genome Annotator Light (GAL)
 https://github.com/computational-genomics-lab/GAL/blob/master/User_Guide_Latest.pdf
https://github.com/computational-genomics-lab/GAL/blob/master/User_Guide_Latest.pdf
Methods
linear multiple view single scale single focus contiguous no abstraction linear parallel arrangement no interconnection segment sparse type point contiguous type point sparse type segment contiguous typeTool
| Access Format | standalone app |
| Supported Files | fasta other |
| License | Creative Commons Attribution License |
| Tool name | Genome Annotator Light (GAL) |
| Tool Link | https://github.com/computational-genomics-lab/GAL |
| Documentation | https://github.com/computational-genomics-lab/GAL/blob/master/User_Guide_Latest.pdf |
Paper
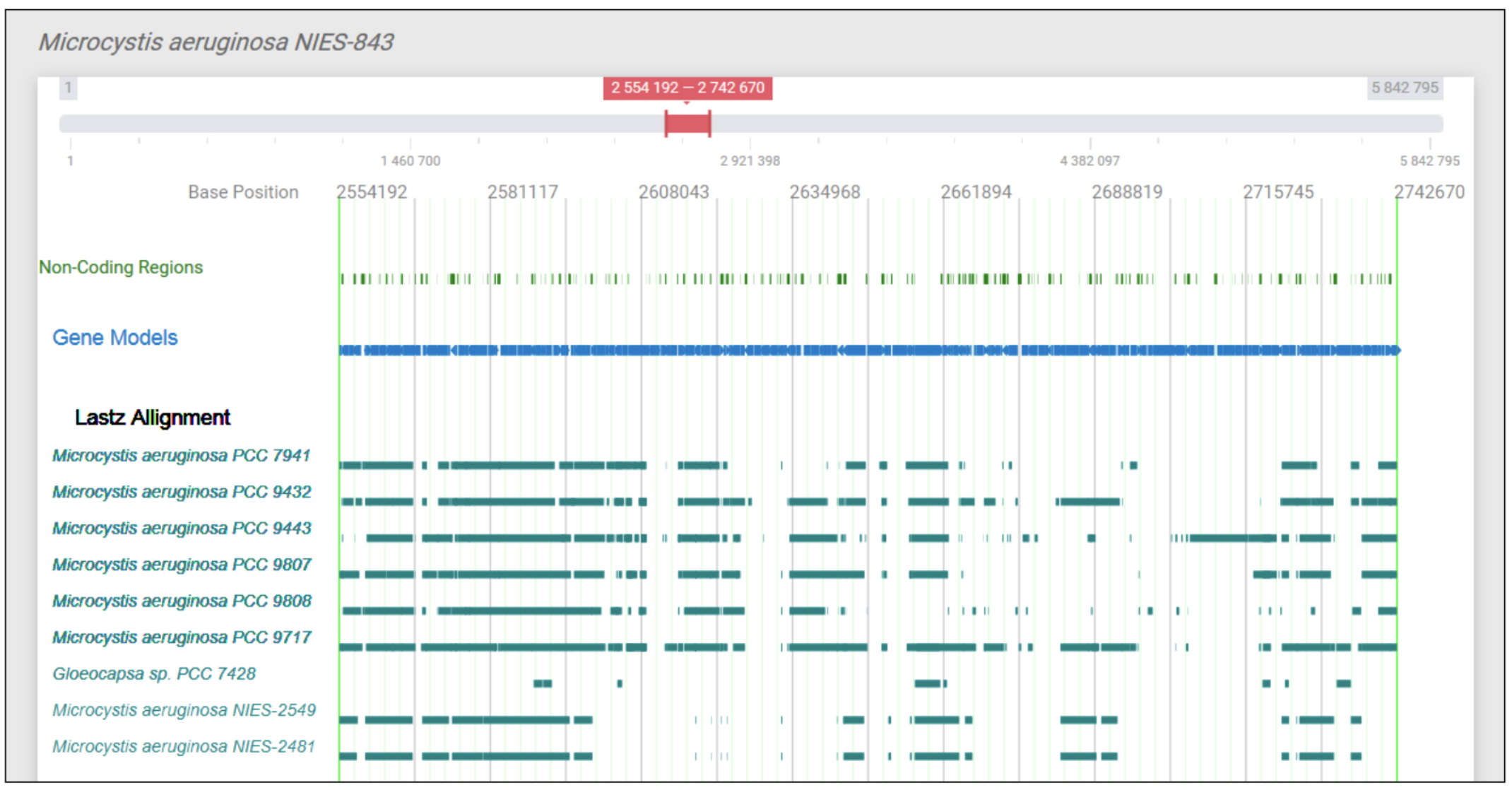
Genome Annotator Light (GAL): A Docker-based package for genome analysis and visualization
Panda, Arijit, Narendrakumar M. Chaudhari, and Sucheta Tripathy. Genome Annotator Light (GAL): A Docker-based package for genome analysis and visualization. Genomics 112.1 (2020): 127-134.
Abstract
Next generation sequencing techniques produce enormous data but its analysis and visualization remains a big challenge. To address this, we have developed Genome Annotator Light(GAL), a Docker based package for genome analysis and data visualization. GAL integrated several existing tools and in-house programs inside a Docker Container for systematic analysis and visualization of genomes through web browser. GAL takes varieties of input types ranging from raw Fasta files to fully annotated files, processes them through a standard annotation pipeline and visualizes on a web browser. Comparative genomic analysis is performed automatically within a given taxonomic class. GAL creates interactive genome browser with clickable genomic feature tracks; local BLAST-able database; query page, on-fly downstream data analysis using EMBOSS etc. Overall, GAL is an extremely convenient, portable and platform independent. Fully integrated web-resources can be easily created and deployed, e.g. www.eumicrobedb.org/cglab, for our in-house genomes. GAL is freely available at https://hub.docker.com/u/cglabiicb/.