CRAMER
 https://github.com/FadyMohareb/cramer#how-to-install-cramer
https://github.com/FadyMohareb/cramer#how-to-install-cramer
Methods
linear multiple view multiple scale multiple focus contiguous no abstraction linear parallel arrangement no interconnection segment sparse typeTool
| Access Format | “programming library” |
| Supported Files | vcf bed other |
| License | MIT |
| Tool name | CRAMER |
| Tool Link | https://github.com/FadyMohareb/cramer |
| Documentation | https://github.com/FadyMohareb/cramer |
Paper
CRAMER: a lightweight, highly customizable web-based genome browser supporting multiple visualization instances
Anastasiadi M, Bragin E, Biojoux P, Ahamed A, Burgin J, de Castro Cogle K, Llaneza-Lago S, Muvunyi R, Scislak M, Aktan I, Molitor C. CRAMER: a lightweight, highly customizable web-based genome browser supporting multiple visualization instances. Bioinformatics. 2020 Jun 1;36(11):3556-7.
Abstract
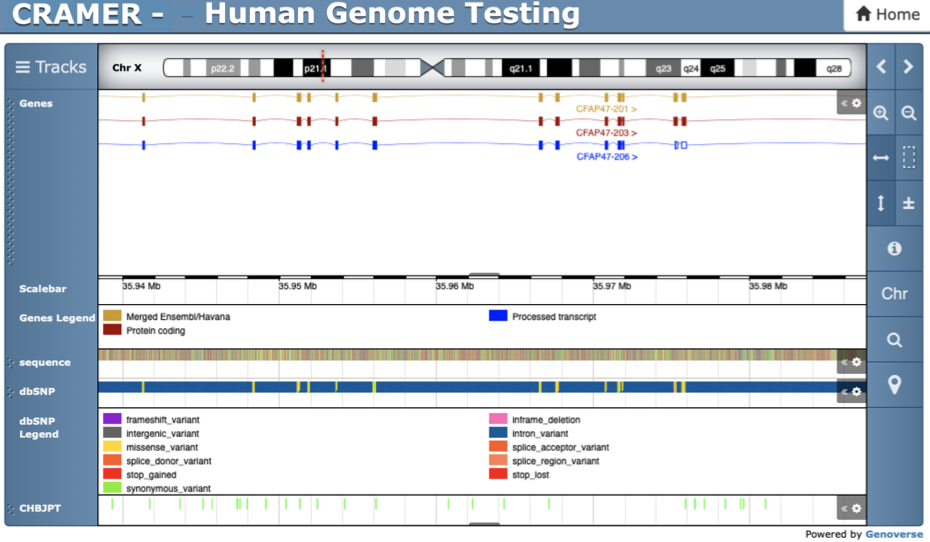
In recent years the ability to generate genomic data has increased dramatically along with the demand for easily personalised and customisable genome browsers for effective visualisation of diverse types of data. Despite the large number of web-based genome browsers available nowadays, none of the existing tools provide means for creating multiple visualisation instances without manual set up on the deployment server side. The Cranfield Genome Browser (CRAMER) is an open-source, lightweight and highly customisable web application for interactive visualisation of genomic data. Once deployed, CRAMER supports seamless creation of multiple visualisation instances in parallel while allowing users to control and customise multiple tracks. The application is deployed on a Node.js server and is supported by a MongoDB database which stored all customisations made by the users allowing quick navigation between instances. Currently, the browser supports visualising a large number of file formats for genome annotation, variant calling, reads coverage and gene expression. Additionally, the browser supports direct Javascript coding for personalised tracks, providing a whole new level of customisation both functionally and visually. Tracks can be added via direct file upload or processed in real-time via links to files stored remotely on an FTP repository. Furthermore, additional tracks can be added by users via simple drag and drop to an existing visualisation instance. Availability and implementation: CRAMER is implemented in JavaScript and is publicly available on GitHub on https://github.com/FadyMohareb/cramer. The application is released under an MIT licence and can be deployed on any server running Linux or Mac OS. Supplementary information: Supplementary data are available at Bioinformatics online.