Xena
 https://xenabrowser.net/?bookmark=501ed28f4548b872967b71b098fe7f65
https://xenabrowser.net/?bookmark=501ed28f4548b872967b71b098fe7f65
Methods
linear multiple view multiple scale multiple focus segregated no abstraction no arrangement no interconnection segment sparse type point sparse typeTool
| Access Format | web application |
| Supported Files | txttab other |
| License | Apache-2.0 |
| Tool name | Xena |
| Tool Link | http://xena.ucsc.edu/ |
| Documentation | https://ucsc-xena.gitbook.io/project/ |
Paper
The UCSC Xena Platform for cancer genomics data visualization and interpretation
Goldman M, Craft B, Brooks AN, Zhu J, Haussler D. The UCSC Xena Platform for cancer genomics data visualization and interpretation [Internet]. bioRxiv. 2018. p. 326470. doi:10.1101/326470
Abstract
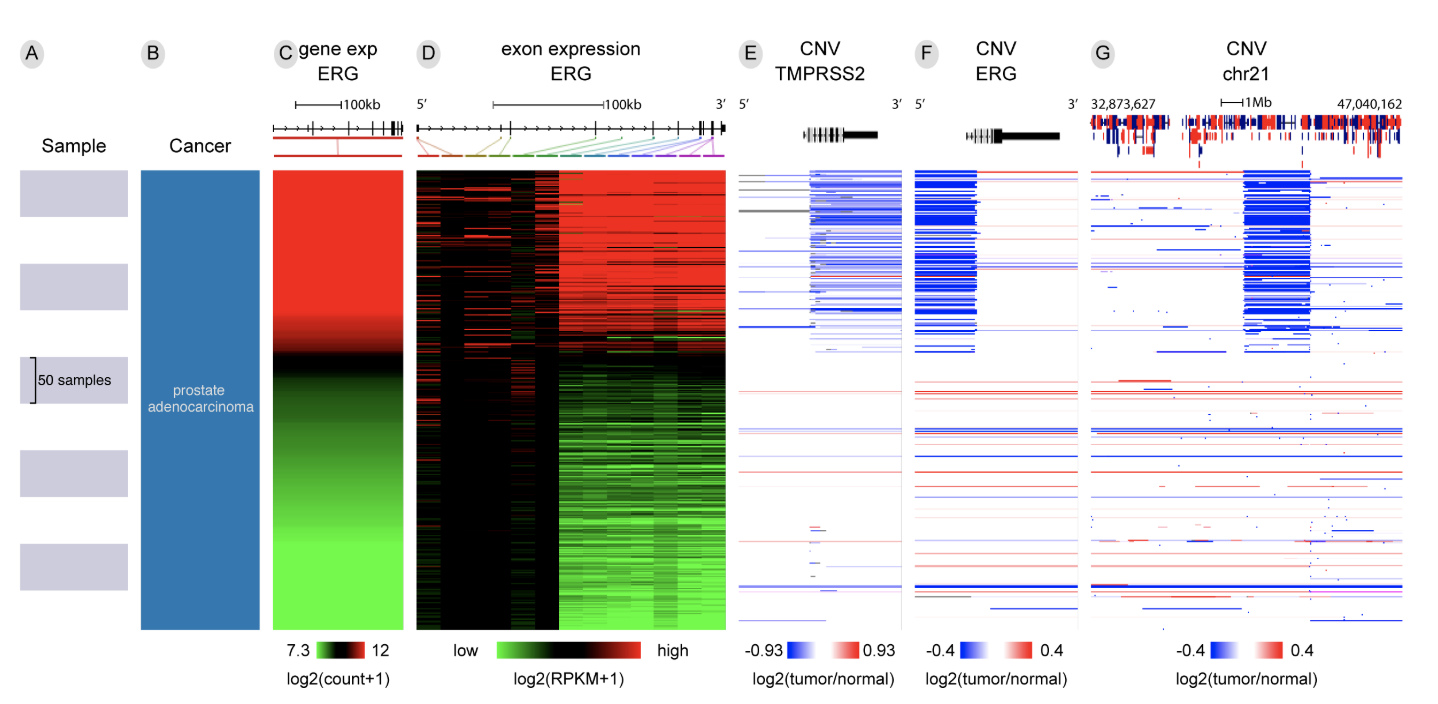
UCSC Xena is a web-based visual integration and exploration tool for multi-omic data and associated clinical and phenotypic annotations. The platform consists of a web-based Xena Browser and turn-key Xena Hubs. Xena showcases seminal cancer genomics datasets from TCGA, Pan-Cancer Atlas, PCAWG, ICGC, and the GDC; a total of more than 1500 datasets across 50 cancer types. We support virtually any functional genomics data modality, including SNVs, INDELs, large structural variants, CNV, gene-and other types of expression, DNA methylation, clinical and phenotypic annotations. A researcher can host their own data securely via a private hub on a laptop or behind a firewall, with visual and analytical integration occurring only within the Xena Browser. Browser features include our high performance Visual Spreadsheet, dynamic Kaplan-Meier survival analysis, powerful filtering and subgrouping, statistical analyses, genomic signatures, bookmarks, box plots, and scatter plots.