VCF Plotein
 https://www.google.com/url?sa=i&url=https%3A%2F%2Fvcfplotein.liigh.unam.mx%2F&psig=AOvVaw3htrc4bxvrx2J7J8ZkHbXp&ust=1605582770728000&source=images&cd=vfe&ved=0CAIQjRxqFwoTCPinlpmMhu0CFQAAAAAdAAAAABAK
https://www.google.com/url?sa=i&url=https%3A%2F%2Fvcfplotein.liigh.unam.mx%2F&psig=AOvVaw3htrc4bxvrx2J7J8ZkHbXp&ust=1605582770728000&source=images&cd=vfe&ved=0CAIQjRxqFwoTCPinlpmMhu0CFQAAAAAdAAAAABAK
Methods
linear single view single scale single focus contiguous no abstraction linear parallel arrangement no interconnection point sparse typeTool
| Access Format | web application |
| Supported Files | vcf |
| License | Creative Commons Attribution License |
| Tool name | VCF Plotein |
| Tool Link | https://vcfplotein.liigh.unam.mx/#/ |
| Documentation | https://vcfplotein.liigh.unam.mx/#/ |
Paper
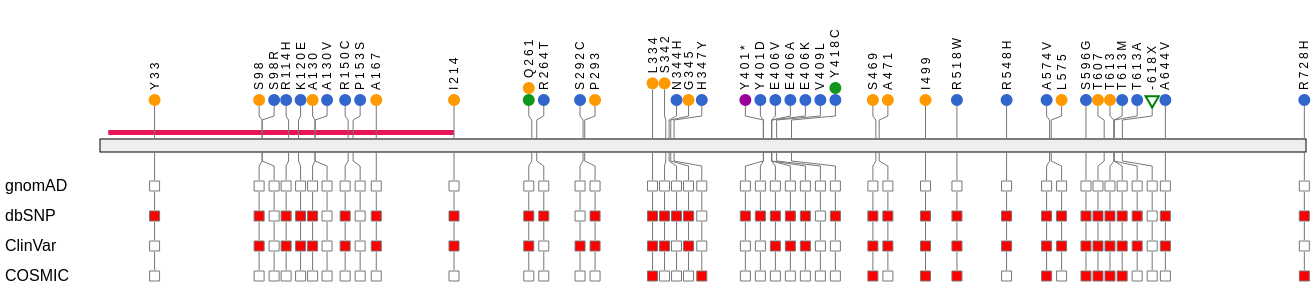
VCF/Plotein: visualization and prioritization of genomic variants from human exome sequencing projects
Raul Ossio, O Isaac Garcia-Salinas, Diego Said Anaya-Mancilla, Jair S Garcia-Sotelo, Luis A Aguilar, David J Adams, Carla Daniela Robles-Espinoza, VCF/Plotein: visualization and prioritization of genomic variants from human exome sequencing projects, Bioinformatics, Volume 35, Issue 22, 15 November 2019, Pages 4803–4805, https://doi.org/10.1093/bioinformatics/btz458
Abstract
Identifying disease-causing variants from exome sequencing projects remains a challenging task that often requires bioinformatics expertise. Here we describe a user-friendly graphical application that allows medical professionals and bench biologists to prioritize and visualize genetic variants from human exome sequencing data. We have implemented VCF/Plotein, a graphical, fully interactive web application able to display exome sequencing data in VCF format. Gene and variant information is extracted from Ensembl. Cross-referencing with external databases and application-based gene and variant filtering have also been implemented. All data processing is done locally by the user’s CPU to ensure the security of patient data.