SynMap2
 https://genomevolution.org/coge/SynMap.pl
https://genomevolution.org/coge/SynMap.pl
Methods
linear single view single scale single focus contiguous no abstraction linear orthogonal arrangement between interconnection segment contiguous typeTool
| Access Format | web application |
| Supported Files | fasta sambamcram other |
| License | MIT |
| Tool name | SynMap2 |
| Tool Link | https://genomevolution.org/coge/SynMap.pl |
| Documentation | https://genomevolution.org/wiki/index.php/Tutorials |
Paper
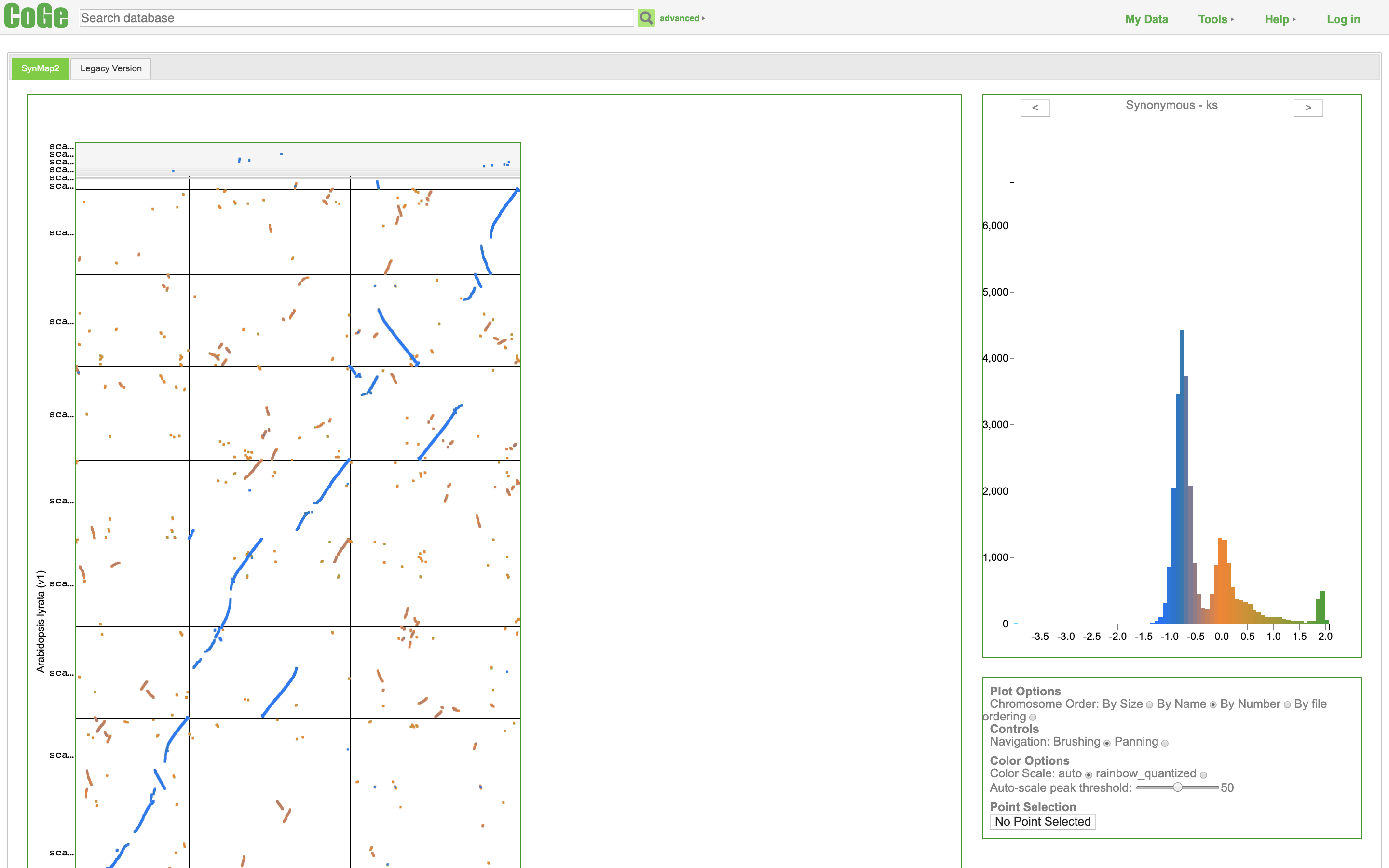
SynMap2 and SynMap3D: web-based whole-genome synteny browsers.
Haug-Baltzell A, Stephens SA, Davey S, Scheidegger CE, Lyons E. SynMap2 and SynMap3D: web-based whole-genome synteny browsers. Bioinformatics. academic.oup.com; 2017;33: 2197–2198.
Abstract
SUMMARY: Current synteny visualization tools either focus on small regions of sequence and do not illustrate genome-wide trends, or are complicated to use and create visualizations that are difficult to interpret. To address this challenge, The Comparative Genomics Platform (CoGe) has developed two web-based tools to visualize synteny across whole genomes. SynMap2 and SynMap3D allow researchers to explore whole genome synteny patterns (across two or three genomes, respectively) in responsive, web-based visualization and virtual reality environments. Both tools have access to the extensive CoGe genome database (containing over 30 000 genomes) as well as the option for users to upload their own data. By leveraging modern web technologies there is no installation required, making the tools widely accessible and easy to use. AVAILABILITY AND IMPLEMENTATION: Both tools are open source (MIT license) and freely available for use online through CoGe ( https://genomevolution.org ). SynMap2 and SynMap3D can be accessed at http://genomevolution.org/coge/SynMap.pl and http://genomevolution.org/coge/SynMap3D.pl , respectively. Source code is available: https://github.com/LyonsLab/coge .