SWAV
 http://swav.popgenetics.net/
http://swav.popgenetics.net/
Methods
linear multiple view single scale single focus contiguous partial abstraction linear parallel arrangement no interconnection segment sparse typeTool
| Access Format | web application |
| Supported Files | fasta other |
| License | MIT Apache 2.0 |
| Tool name | SWAV |
| Tool Link | http://swav.popgenetics.net/ |
| Documentation | http://swav.popgenetics.net/ |
Paper
SWAV: a web-based visualization browser for sliding window analysis
Zhu, Z., Wang, Y., Zhou, X. et al. SWAV: a web-based visualization browser for sliding window analysis. Sci Rep 10, 149 (2020). https://doi.org/10.1038/s41598-019-57038-x
Abstract
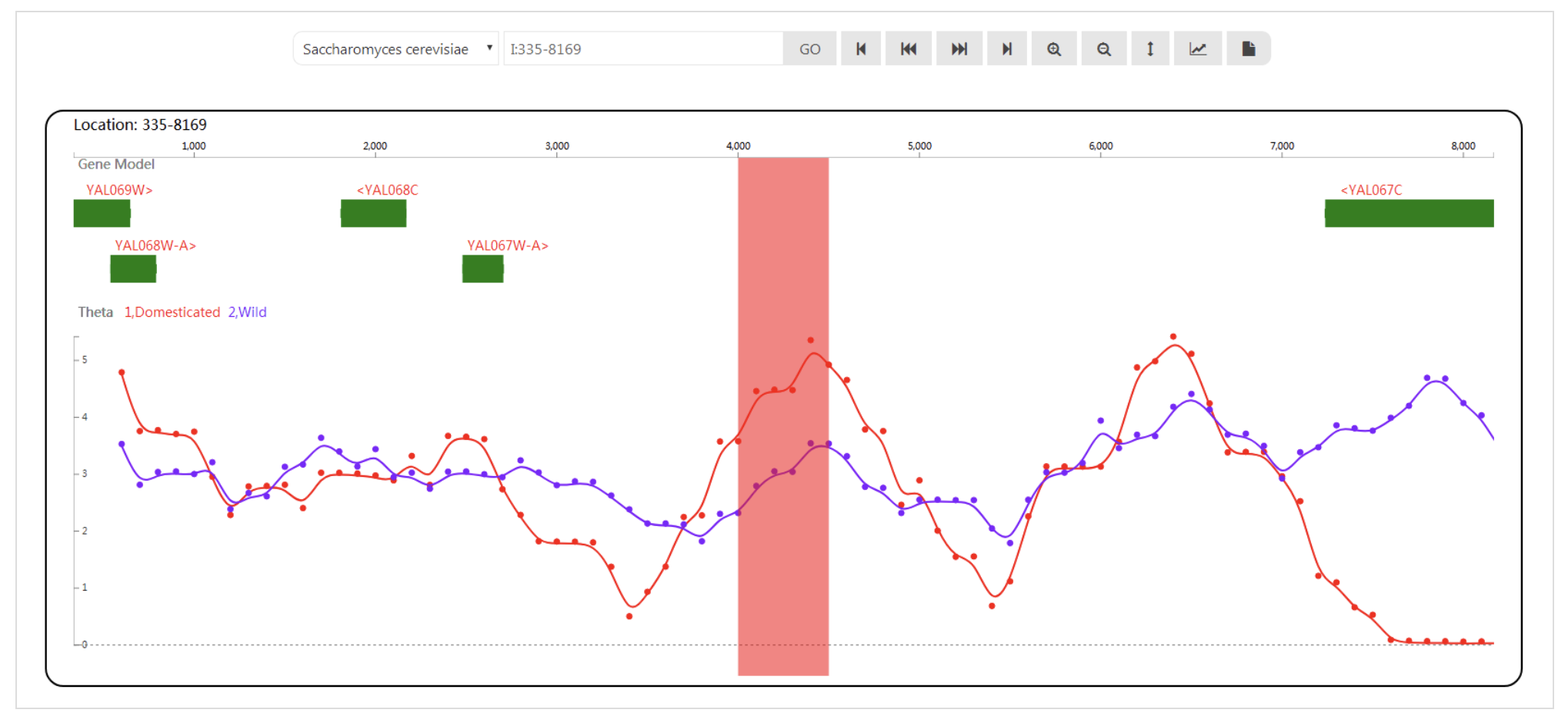
Sliding window analysis has been extensively applied in evolutionary biology. With the development of the high-throughput DNA sequencing of organisms at the population level, an application that is dedicated to visualizing population genetic test statistics at the genomic level is needed. We have developed the sliding window analysis viewer (SWAV), which is a web-based program that can be used to integrate, view and browse test statistics and perform genome annotation. In addition to browsing, SAV can mark, generate and customize statistical images and search by sequence alignment, position or gene name. These features facilitate the effectiveness of sliding window analysis. As an example application, yeast and silkworm resequencing data are analyzed with SWAV. The SWAV package, user manual and usage demo are available at http://swav.popgenetics.net.