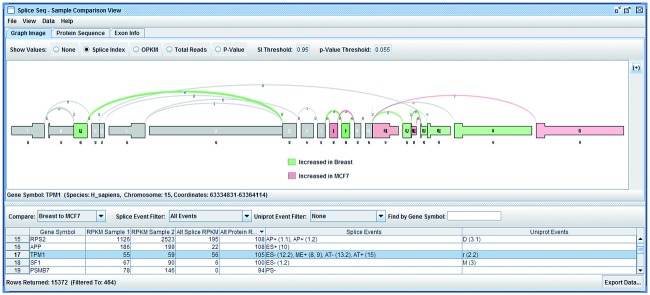
SpliceSeq
 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3436850/figure/bts452-F1/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3436850/figure/bts452-F1/
Methods
linear single view single scale single focus segregated partial abstraction no arrangement no interconnection segment sparse typeTool
| Access Format | standalone app |
| Supported Files | fasta other |
| License | open source |
| Tool name | SpliceSeq |
| Tool Link | https://bioinformatics.mdanderson.org/public-software/spliceseq/ |
| Documentation | https://bioinformatics.mdanderson.org/public-software/spliceseq/installation/ |
Paper
SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts.
Ryan MC, Cleland J, Kim R, Wong WC, Weinstein JN. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics. academic.oup.com; 2012;28: 2385–2387.
Abstract
SpliceSeq is a resource for RNA-Seq data that provides a clear view of alternative splicing and identifies potential functional changes that result from splice variation. It displays intuitive visualizations and prioritized lists of results that highlight splicing events and their biological consequences. SpliceSeq unambiguously aligns reads to gene splice graphs, facilitating accurate analysis of large, complex transcript variants that cannot be adequately represented in other formats.