Sashimi Plot
 https://software.broadinstitute.org/software/igv/Sashimi
https://software.broadinstitute.org/software/igv/Sashimi
Methods
linear single view single scale single focus contiguous no abstraction no arrangement within interconnection segment sparse type point sparse typeTool
| Access Format | web application |
| Supported Files | bed fasta sambamcram vcf other |
| License | MIT |
| Tool name | Sashimi Plot |
| Tool Link | https://software.broadinstitute.org/software/igv/Sashimi |
| Documentation | https://software.broadinstitute.org/software/igv/Sashimi |
Paper
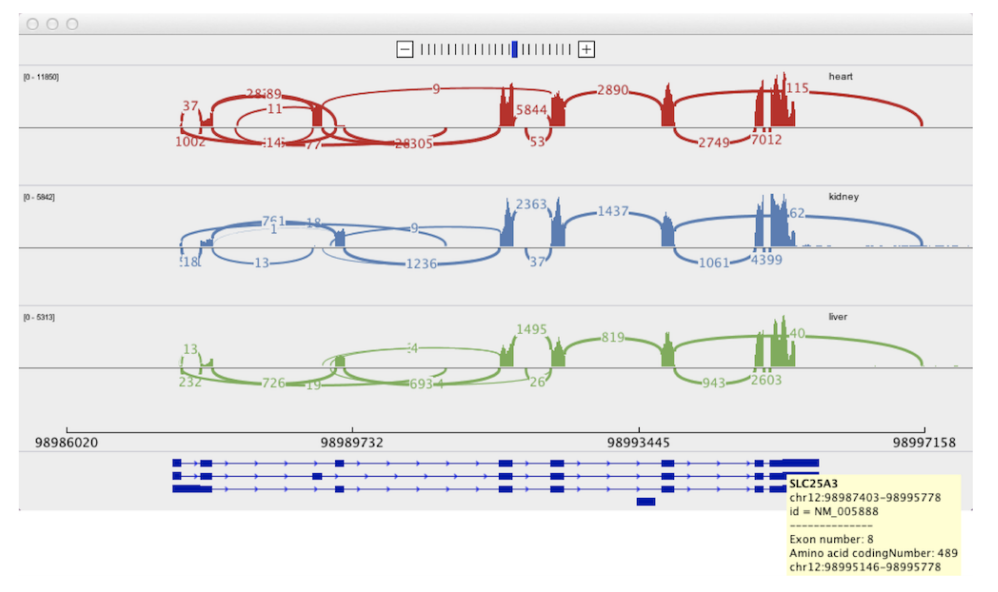
Sashimi plots: Quantitative visualization of RNA sequencing read alignments
Katz Y, Wang ET, Silterra J, Schwartz S, Wong B, Mesirov JP, et al. Sashimi plots: Quantitative visualization of RNA sequencing read alignments [Internet]. arXiv [q-bio.GN]. 2013. Available: http://arxiv.org/abs/1306.3466
Abstract
Motivation: Analysis of RNA sequencing (RNA-Seq) data revealed that the vast majority of human genes express multiple mRNA isoforms, produced by alternative pre-mRNA splicing and other mechanisms, and that most alternative isoforms vary in expression between human tissues. As RNA-Seq datasets grow in size, it remains challenging to visualize isoform expression across multiple samples. Results: To help address this problem, we present Sashimi plots, a quantitative visualization of aligned RNA-Seq reads that enables quantitative comparison of exon usage across samples or experimental conditions. Sashimi plots can be made using the Broad Integrated Genome Viewer or with a stand-alone command line program.