Rondo
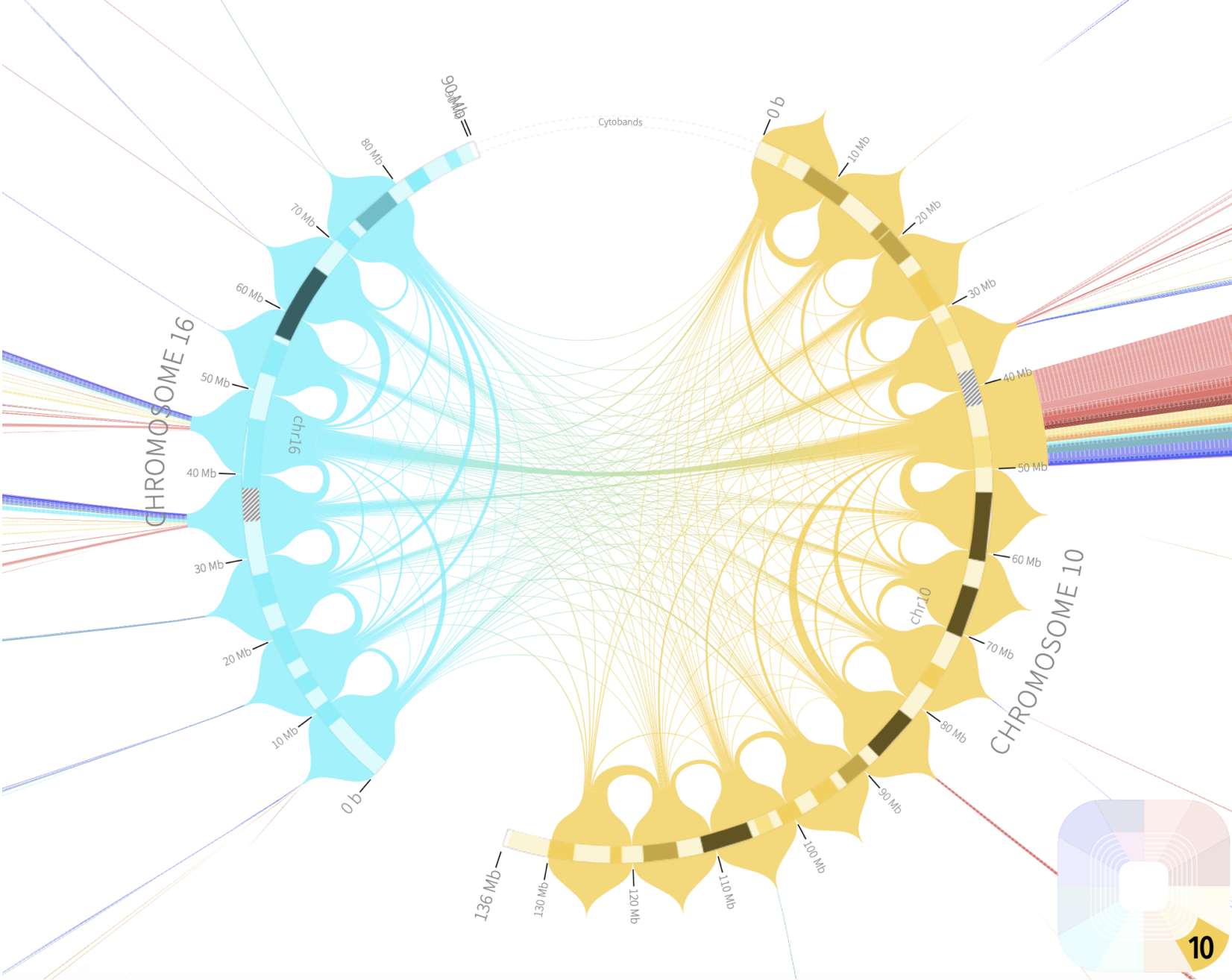 http://rondo.ws/
http://rondo.ws/
Methods
circular single view single scale single focus contiguous no abstraction no arrangement within interconnection segment sparse type segment contiguous type point sparse type point contiguous typeTool
| Access Format | web application |
| Supported Files | notapplicable |
| License | unavailable |
| Tool name | Rondo |
| Tool Link | http://www.rondo.ws/ |
| Documentation | https://www.youtube.com/watch?v=s8eorV5aL6U&feature=youtu.be |
Paper
Three-dimensional disorganization of the cancer genome occurs coincident with long-range genetic and epigenetic alterations.
Taberlay PC, Achinger-Kawecka J, Lun ATL, Buske FA, Sabir K, Gould CM, et al. Three-dimensional disorganisation of the cancer genome occurs coincident with long range genetic and epigenetic alterations. Genome Res. genome.cshlp.org; 2016; doi:10.1101/gr.201517.115
Abstract
A three-dimensional chromatin state underpins the structural and functional basis of the genome by bringing regulatory elements and genes into close spatial proximity to ensure proper, cell-type-specific gene expression profiles. Here, we performed Hi-C chromosome conformation capture sequencing to investigate how three-dimensional chromatin organization is disrupted in the context of copy-number variation, long-range epigenetic remodeling, and atypical gene expression programs in prostate cancer. We find that cancer cells retain the ability to segment their genomes into megabase-sized topologically associated domains (TADs); however, these domains are generally smaller due to establishment of additional domain boundaries. Interestingly, a large proportion of the new cancer-specific domain boundaries occur at regions that display copy-number variation. Notably, a common deletion on 17p13.1 in prostate cancer spanning the TP53 tumor suppressor locus results in bifurcation of a single TAD into two distinct smaller TADs. Change in domain structure is also accompanied by novel cancer-specific chromatin interactions within the TADs that are enriched at regulatory elements such as enhancers, promoters, and insulators, and associated with alterations in gene expression. We also show that differential chromatin interactions across regulatory regions occur within long-range epigenetically activated or silenced regions of concordant gene activation or repression in prostate cancer. Finally, we present a novel visualization tool that enables integrated exploration of Hi-C interaction data, the transcriptome, and epigenome. This study provides new insights into the relationship between long-range epigenetic and genomic dysregulation and changes in higher-order chromatin interactions in cancer.