PSU 3D Genome Browser
 https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1519-9
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1519-9
Methods
linear single view single scale single focus segregated no abstraction no arrangement within interconnection between interconnection segment sparse type segment contiguous type point sparse type point contiguous typeTool
| Access Format | web application |
| Supported Files | other |
| License | unavailable |
| Tool name | PSU 3D Genome Browser |
| Tool Link | http://3dgenome.org |
| Documentation | http://promoter.bx.psu.edu/hi-c/tutorial.html |
Paper
The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions
Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. genomebiology.biomedcentral.com; 2018;19: 151.
Abstract
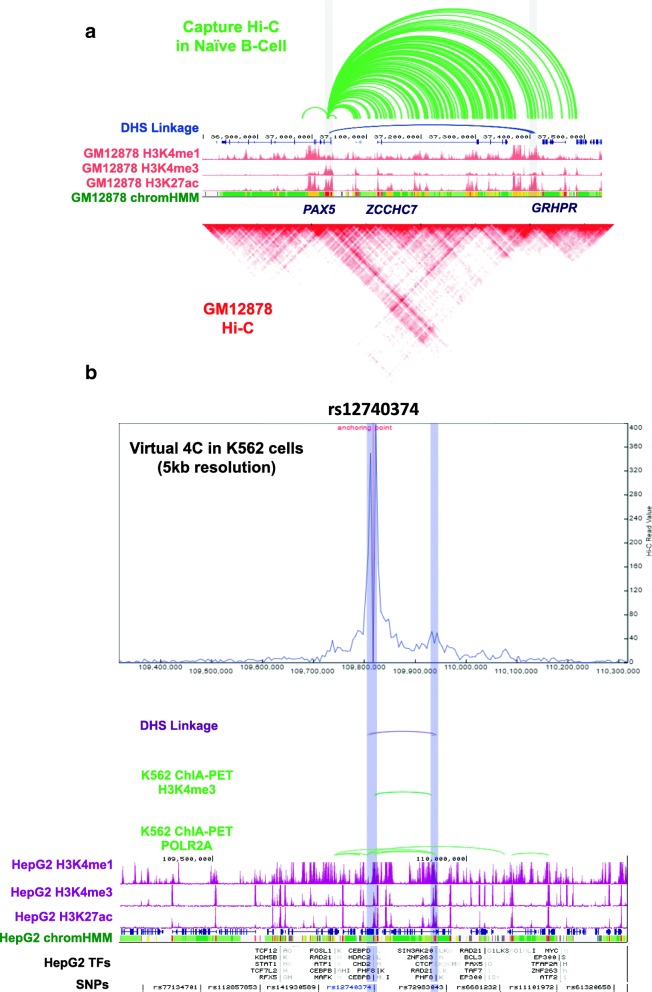
Here, we introduce the 3D Genome Browser, http://3dgenome.org , which allows users to conveniently explore both their own and over 300 publicly available chromatin interaction data of different types. We design a new binary data format for Hi-C data that reduces the file size by at least a magnitude and allows users to visualize chromatin interactions over millions of base pairs within seconds. Our browser provides multiple methods linking distal cis-regulatory elements with their potential target genes. Users can seamlessly integrate thousands of other omics data to gain a comprehensive view of both regulatory landscape and 3D genome structure.