Oviz-Bio
 https://academic.oup.com/view-large/figure/205119849/gkaa371fig1.jpg
https://academic.oup.com/view-large/figure/205119849/gkaa371fig1.jpg
Methods
linear multiple view single scale single focus contiguous no abstraction linear parallel arrangement no interconnection segment contiguous type segment sparse type point contiguous type point sparse typeTool
| Access Format | web application |
| Supported Files | other |
| License | Creative Commons Attribution License |
| Tool name | Oviz-Bio |
| Tool Link | https://bio.oviz.org/ |
| Documentation | https://bio.oviz.org/demo-project/analyses/genome-browser |
Paper
Oviz-Bio: a web-based platform for interactive cancer genomics data visualization
Wenlong Jia, Hechen Li, Shiying Li, Lingxi Chen, Shuai Cheng Li, Oviz-Bio: a web-based platform for interactive cancer genomics data visualization, Nucleic Acids Research, Volume 48, Issue W1, 02 July 2020, Pages W415–W426, https://doi.org/10.1093/nar/gkaa371
Abstract
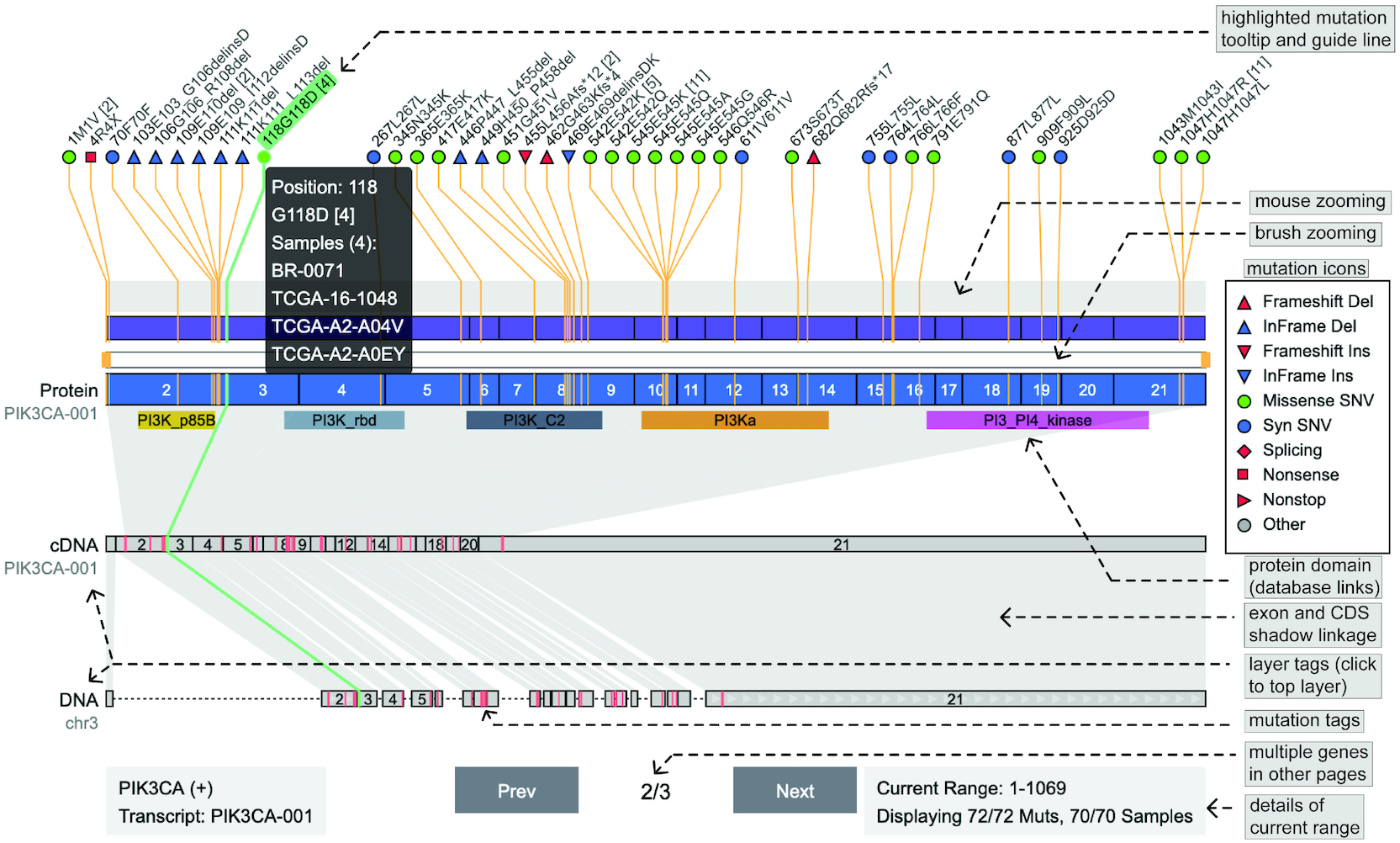
Genetics data visualization plays an important role in the sharing of knowledge from cancer genome research. Many types of visualization are widely used, most of which are static and require sufficient coding experience to create. Here, we present Oviz-Bio, a web-based platform that provides interactive and real-time visualizations of cancer genomics data. Researchers can interactively explore visual outputs and export high-quality diagrams. Oviz-Bio supports a diverse range of visualizations on common cancer mutation types, including annotation and signatures of small scale mutations, haplotype view and focal clusters of copy number variations, split-reads alignment and heatmap view of structural variations, transcript junction of fusion genes and genomic hotspot of oncovirus integrations. Furthermore, Oviz-Bio allows landscape view to investigate multi-layered data in samples cohort. All Oviz-Bio visual applications are freely available at https://bio.oviz.org/.