MochiView
 http://www.johnsonlab.ucsf.edu/mochi
http://www.johnsonlab.ucsf.edu/mochi
Methods
linear multiple view multiple scale single focus segregated no abstraction no arrangement no interconnection segment sparse type point sparse type point contiguous typeTool
| Access Format | standalone app |
| Supported Files | fasta bed other |
| License | GNU Library/Lesser General Public License (LGPL) |
| Tool name | MochiView |
| Tool Link | http://www.johnsonlab.ucsf.edu/mochi |
| Documentation | https://static1.squarespace.com/static/56463117e4b0770d2cd2d163/t/564bbbb1e4b0c03db1589e9a/1447803825330/MochiView_Manual_v1.45.pdf |
Paper
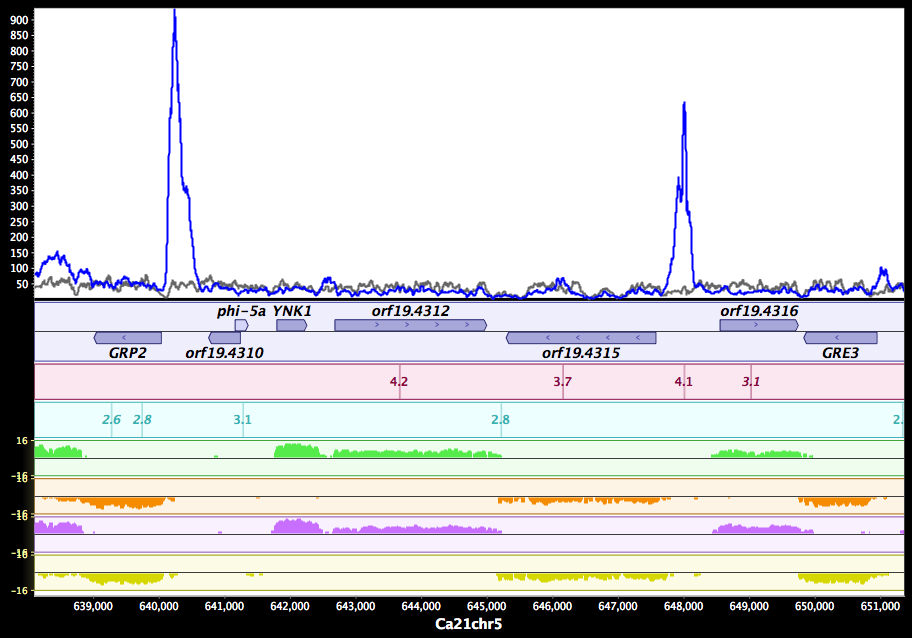
MochiView: versatile software for genome browsing and DNA motif analysis
Homann OR, Johnson AD. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol. bmcbiol.biomedcentral.com; 2010;8: 49.
Abstract
As high-throughput technologies rapidly generate genome-scale data, it becomes increasingly important to visually integrate these data so that specific hypotheses can be formulated and tested., , We present MochiView, a platform-independent Java software that integrates browsing of genomic sequences, features, and data with DNA motif visualization and analysis in a visually-appealing and user-friendly application., , While highly versatile, the software is particularly useful for organizing, exploring, and analyzing large genomic data sets, such as those from deep RNA sequencing, chromatin immunoprecipitation experiments (ChIP-Seq and ChIP-Chip), and transcriptional profiling. MochiView provides an extensive suite of utilities to identify and to explore connections between these data sets and short sequence motifs present in DNA or RNA.