MicroScope
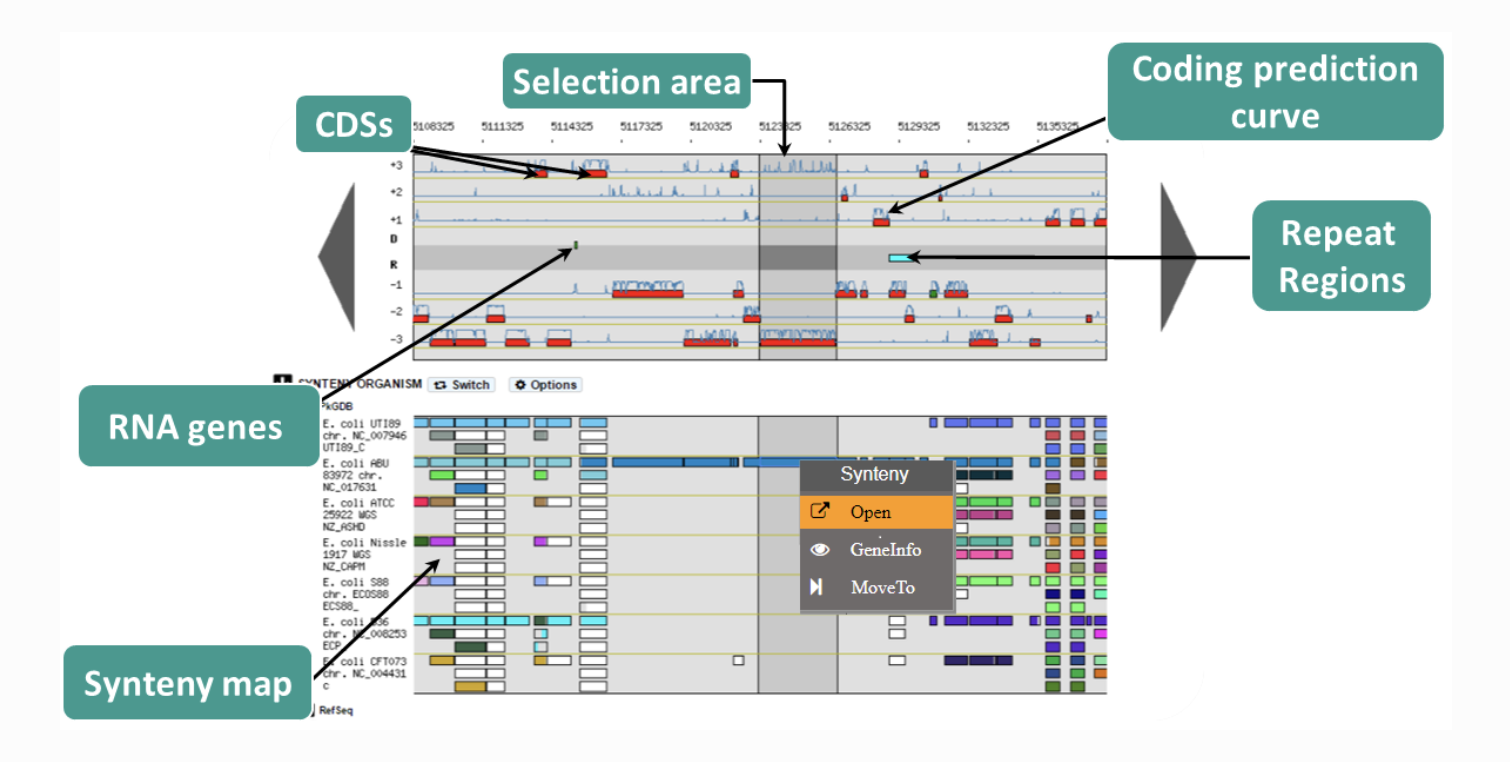 https://microscope.readthedocs.io/en/v3.14.1/content/mage/viewer.html
https://microscope.readthedocs.io/en/v3.14.1/content/mage/viewer.html
Methods
linear circular multiple view single scale single focus contiguous no abstraction linear parallel arrangement between interconnection segment sparse type point contiguous type point sparse type segment contiguous typeTool
| Access Format | web application |
| Supported Files | other |
| License | Creative Commons Attribution License |
| Tool name | MicroScope |
| Tool Link | https://mage.genoscope.cns.fr/microscope/home/index.php |
| Documentation | https://microscope.readthedocs.io/en/v3.14.1/index.html |
Paper
MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis
David Vallenet, Alexandra Calteau, Mathieu Dubois, Paul Amours, Adelme Bazin, Mylène Beuvin, Laura Burlot, Xavier Bussell, Stéphanie Fouteau, Guillaume Gautreau, Aurélie Lajus, Jordan Langlois, Rémi Planel, David Roche, Johan Rollin, Zoe Rouy, Valentin Sabatet, Claudine Médigue, MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis, Nucleic Acids Research, Volume 48, Issue D1, 08 January 2020, Pages D579–D589, https://doi.org/10.1093/nar/gkz926
Abstract
Large-scale genome sequencing and the increasingly massive use of high-throughput approaches produce a vast amount of new information that completely transforms our understanding of thousands of microbial species. However, despite the development of powerful bioinformatics approaches, full interpretation of the content of these genomes remains a difficult task. Launched in 2005, the MicroScope platform (https://www.genoscope.cns.fr/agc/microscope) has been under continuous development and provides analysis for prokaryotic genome projects together with metabolic network reconstruction and post-genomic experiments allowing users to improve the understanding of gene functions. Here we present new improvements of the MicroScope user interface for genome selection, navigation and expert gene annotation. Automatic functional annotation procedures of the platform have also been updated and we added several new tools for the functional annotation of genes and genomic regions. We finally focus on new tools and pipeline developed to perform comparative analyses on hundreds of genomes based on pangenome graphs. To date, MicroScope contains data for >11 800 microbial genomes, part of which are manually curated and maintained by microbiologists (>4500 personal accounts in September 2019). The platform enables collaborative work in a rich comparative genomic context and improves community-based curation efforts.