MGcV
 https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-14-209
https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-14-209
Methods
linear single view single scale single focus contiguous no abstraction no arrangement no interconnection segment sparse typeTool
| Access Format | web application |
| Supported Files | other |
| License | unavailable |
| Tool name | MGcV |
| Tool Link | https://mgcv.cmbi.umcn.nl/ |
| Documentation | https://mgcv.cmbi.umcn.nl/help.html#data_input |
Paper
MGcV: the microbial genomic context viewer for comparative genome analysis
Overmars L, Kerkhoven R, Siezen RJ, Francke C. MGcV: the microbial genomic context viewer for comparative genome analysis. BMC Genomics. bmcgenomics.biomedcentral.com; 2013;14: 209.
Abstract
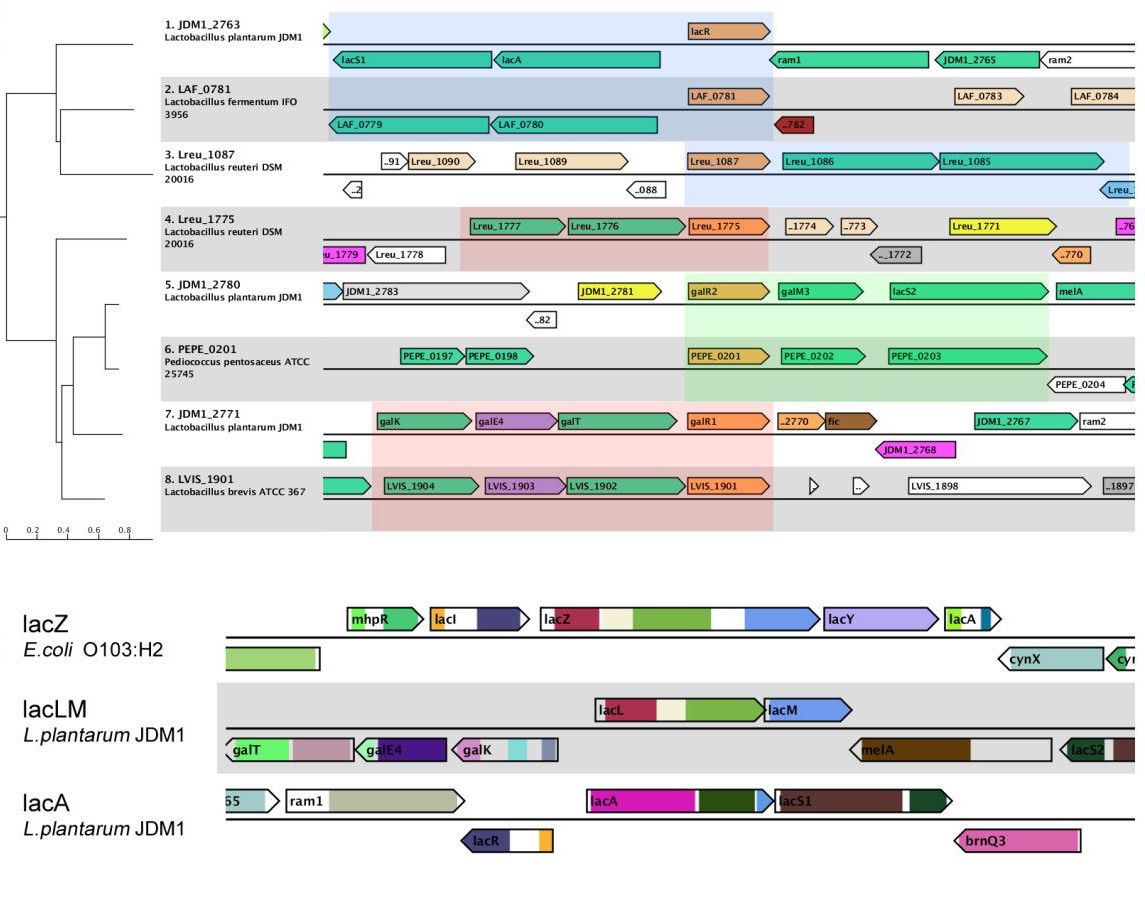
Background: , Conserved gene context is used in many types of comparative genome analyses. It is used to provide leads on gene function, to guide the discovery of regulatory sequences, but also to aid in the reconstruction of metabolic networks. We present the Microbial Genomic context Viewer (MGcV), an interactive, web-based application tailored to strengthen the practice of manual comparative genome context analysis for bacteria., , Results: , MGcV is a versatile, easy-to-use tool that renders a visualization of the genomic context of any set of selected genes, genes within a phylogenetic tree, genomic segments, or regulatory elements. It is tailored to facilitate laborious tasks such as the interactive annotation of gene function, the discovery of regulatory elements, or the sequence-based reconstruction of gene regulatory networks. We illustrate that MGcV can be used in gene function annotation by visually integrating information on prokaryotic genes, like their annotation as available from NCBI with other annotation data such as Pfam domains, sub-cellular location predictions and gene-sequence characteristics such as GC content. We also illustrate the usefulness of the interactive features that allow the graphical selection of genes to facilitate data gathering (e.g. upstream regions, ID’s or annotation), in the analysis and reconstruction of transcription regulation. Moreover, putative regulatory elements and their corresponding scores or data from RNA-seq and microarray experiments can be uploaded, visualized and interpreted in (ranked-) comparative context maps. The ranked maps allow the interpretation of predicted regulatory elements and experimental data in light of each other., , Conclusion: , MGcV advances the manual comparative analysis of genes and regulatory elements by providing fast and flexible integration of gene related data combined with straightforward data retrieval. MGcV is available at http://mgcv.cmbi.ru.nl.