MEXPRESS
 https://mexpress.be/index.html
https://mexpress.be/index.html
Methods
linear single view single scale single focus contiguous no abstraction no arrangement between interconnection segment sparse type segment contiguous type point sparse type point contiguous typeTool
| Access Format | web application |
| Supported Files | notapplicable |
| License | MIT |
| Tool name | MEXPRESS |
| Tool Link | https://mexpress.be/ |
| Documentation | https://mexpress.be/about.html |
Paper
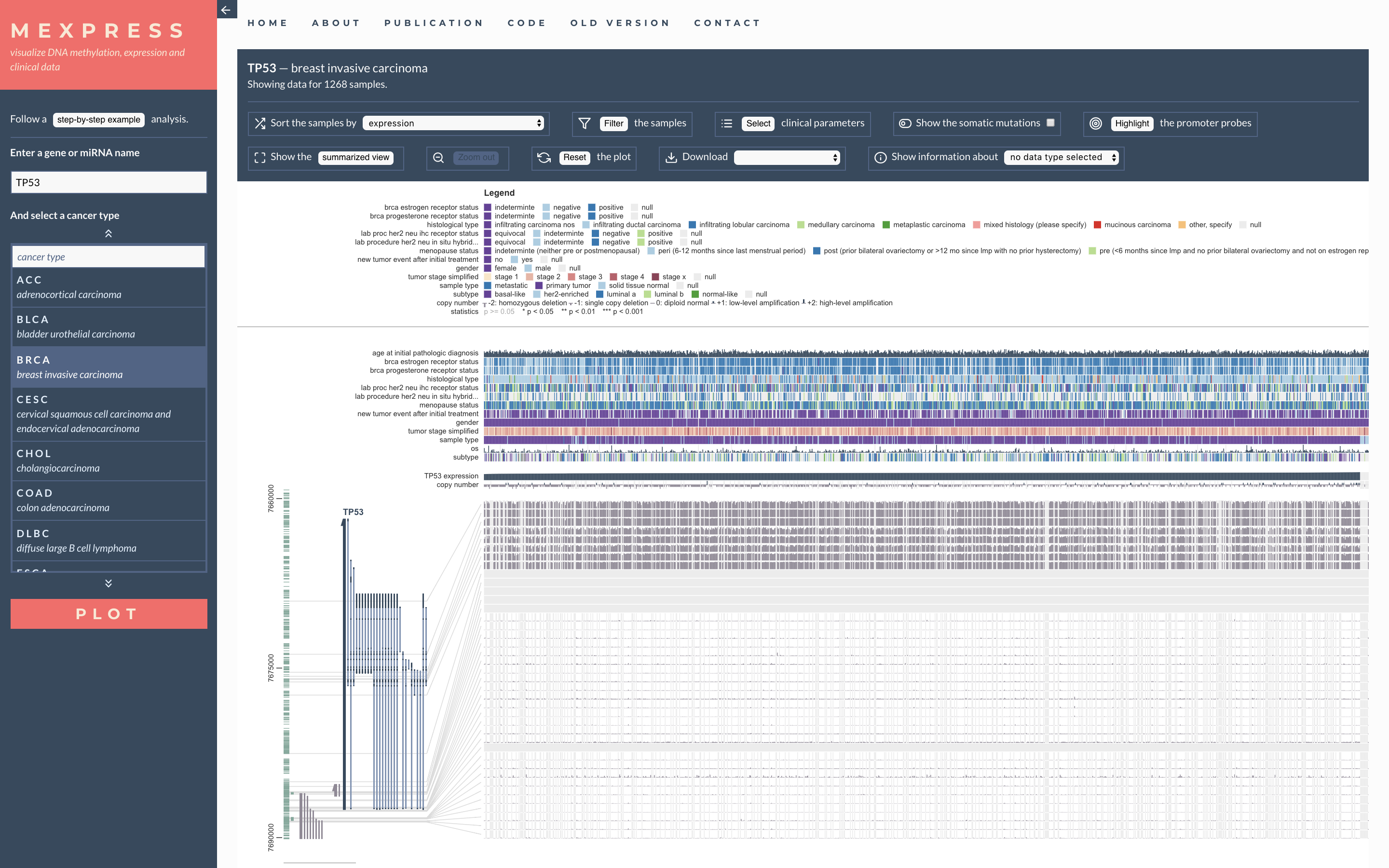
MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data
Koch, A., De Meyer, T., Jeschke, J. and Van Criekinge, W., 2015. MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC genomics, 16(1), p.636.
Abstract
Background: In recent years, increasing amounts of genomic and clinical cancer data have become publically available through large-scale collaborative projects such as The Cancer Genome Atlas (TCGA). However, as long as these datasets are difficult to access and interpret, they are essentially useless for a major part of the research community and their scientific potential will not be fully realized. To address these issues we developed MEXPRESS, a straightforward and easy-to-use web tool for the integration and visualization of the expression, DNA methylation and clinical TCGA data on a single-gene level (http://mexpress.be). Results: In comparison to existing tools, MEXPRESS allows researchers to quickly visualize and interpret the different TCGA datasets and their relationships for a single gene, as demonstrated for GSTP1 in prostate adenocarcinoma. We also used MEXPRESS to reveal the differences in the DNA methylation status of the PAM50 marker gene MLPH between the breast cancer subtypes and how these differences were linked to the expression of MPLH. Conclusions: We have created a user-friendly tool for the visualization and interpretation of TCGA data, offering clinical researchers a simple way to evaluate the TCGA data for their genes or candidate biomarkers of interest.