JBrowse
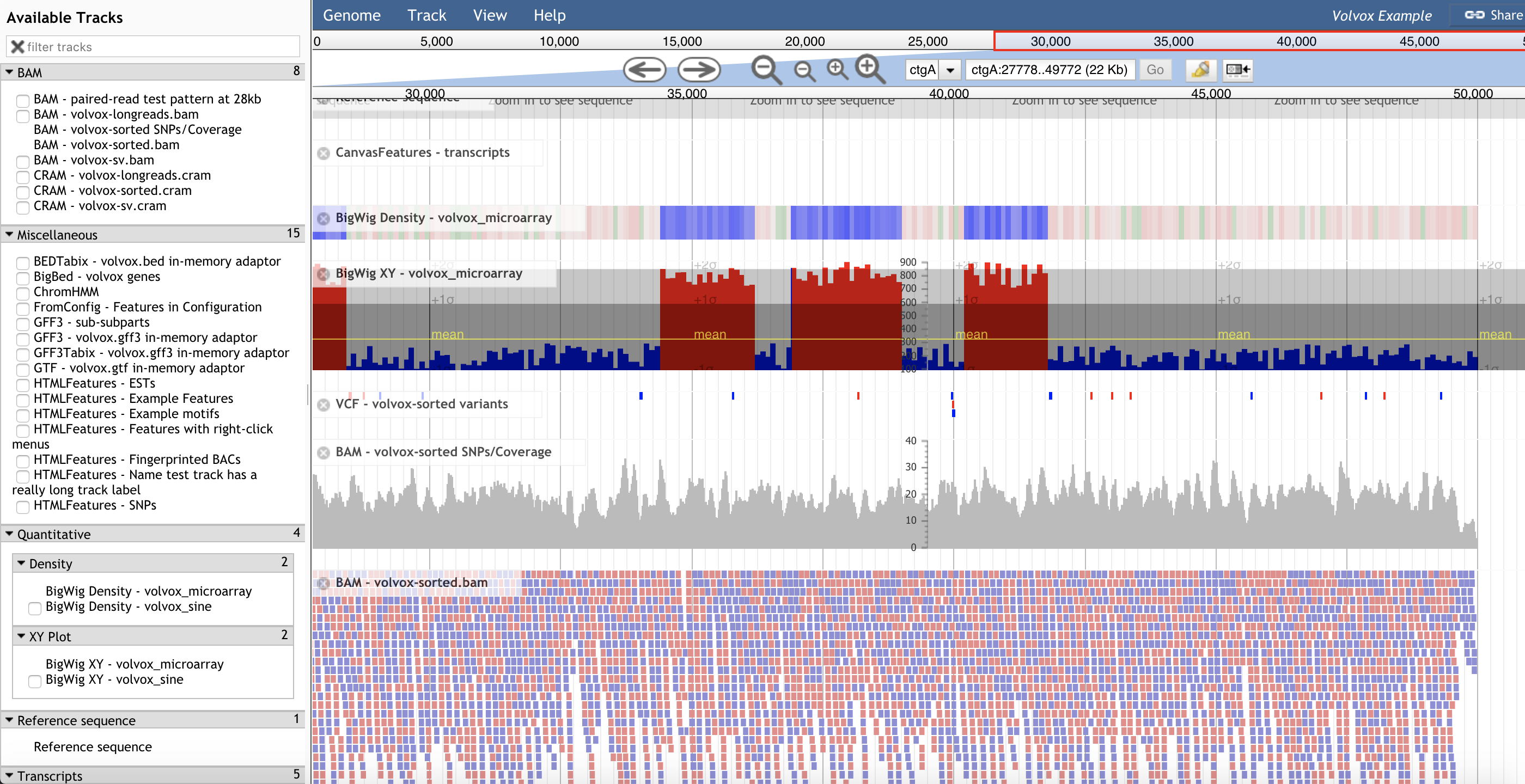 https://jbrowse.org/
https://jbrowse.org/
Methods
linear single view single scale single focus segregated no abstraction no arrangement no interconnection segment sparse type segment contiguous type point sparse type point contiguous typeTool
| Access Format | web application standalone app |
| Supported Files | bed fasta sambamcram vcf other |
| License | GNU LGPL, Artistic License |
| Tool name | JBrowse |
| Tool Link | https://jbrowse.org/ |
| Documentation | https://jbrowse.org/docs/installation.html |
Paper
JBrowse: a dynamic web platform for genome visualization and analysis
Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. genomebiology.biomedcentral.com; 2016;17: 66.
Abstract
Background: JBrowse is a fast and full-featured genome browser built with JavaScript and HTML5. It is easily embedded into websites or apps but can also be served as a standalone web page. Results: Overall improvements to speed and scalability are accompanied by specific enhancements that support complex interactive queries on large track sets. Analysis functions can readily be added using the plugin framework; most visual aspects of tracks can also be customized, along with clicks, mouseovers, menus, and popup boxes. JBrowse can also be used to browse local annotation files offline and to generate high-resolution figures for publication. Conclusions: JBrowse is a mature web application suitable for genome visualization and analysis.