HUGIn
 https://yunliweb.its.unc.edu/HUGIn/#plots
https://yunliweb.its.unc.edu/HUGIn/#plots
Methods
linear single view single scale single focus segregated no abstraction no arrangement between interconnection segment sparse type segment contiguous type point sparse type point contiguous typeTool
| Access Format | web application |
| Supported Files | notapplicable |
| License | unavailable |
| Tool name | HUGIn |
| Tool Link | http://yunliweb.its.unc.edu/HUGIn |
| Documentation | https://yunliweb.its.unc.edu/HUGIn/tutorial_page.php |
Paper
HUGIn: Hi-C Unifying Genomic Interrogator.
Martin JS, Xu Z, Reiner AP, Mohlke KL, Sullivan P, Ren B, et al. HUGIn: Hi-C Unifying Genomic Interrogator. Bioinformatics. academic.oup.com; 2017;33: 3793–3795.
Abstract
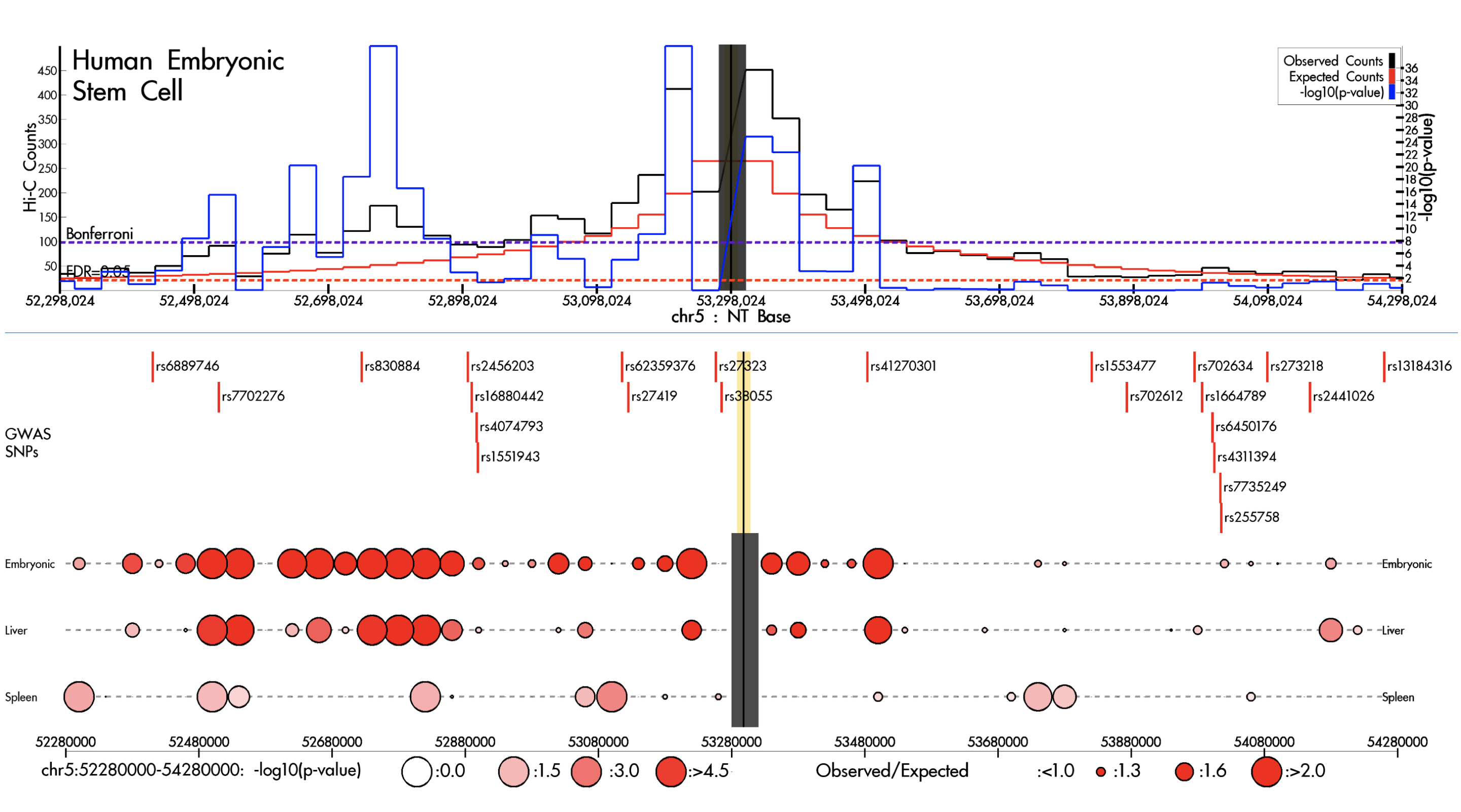
MOTIVATION: High throughput chromatin conformation capture (3C) technologies, such as Hi-C and ChIA-PET, have the potential to elucidate the functional roles of non-coding variants. However, most of published genome-wide unbiased chromatin organization studies have used cultured cell lines, limiting their generalizability. RESULTS: We developed a web browser, HUGIn, to visualize Hi-C data generated from 21 human primary tissues and cell lines. HUGIn enables assessment of chromatin contacts both constitutive across and specific to tissue(s) and/or cell line(s) at any genomic loci, including GWAS SNPs, eQTLs and cis-regulatory elements, facilitating the understanding of both GWAS and eQTL results and functional genomics data. AVAILABILITY AND IMPLEMENTATION: HUGIn is available at http://yunliweb.its.unc.edu/HUGIn.