ggBio
 https://media.springernature.com/full/springer-static/image/art%3A10.1186%2Fgb-2012-13-8-r77/MediaObjects/13059_2012_Article_3010_Fig7_HTML.jpg
https://media.springernature.com/full/springer-static/image/art%3A10.1186%2Fgb-2012-13-8-r77/MediaObjects/13059_2012_Article_3010_Fig7_HTML.jpg
Methods
linear circular single view multiple view single scale single focus segregated contiguous partial abstraction no arrangement within interconnection between interconnection segment sparse type segment contiguous type point sparse type point contiguous typeTool
| Access Format | programming library |
| Supported Files | other |
| License | Artistic-2.0 |
| Tool name | ggBio |
| Tool Link | https://bioconductor.org/packages/release/bioc/html/ggbio.html |
| Documentation | https://bioconductor.org/packages/release/bioc/vignettes/ggbio/inst/doc/ggbio.pdf |
Paper
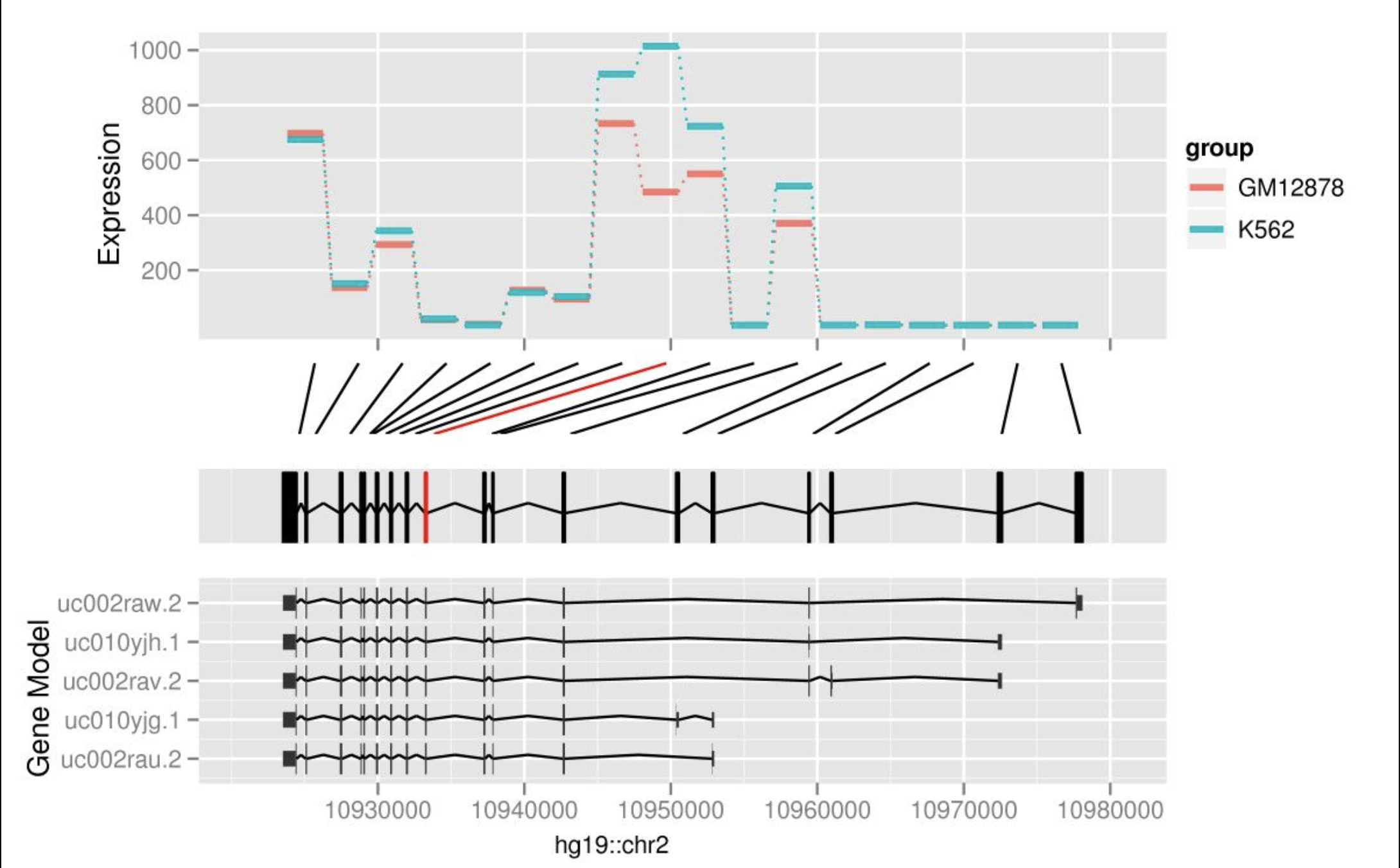
ggbio: an R package for extending the grammar of graphics for genomic data
Yin T, Cook D, Lawrence M. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. genomebiology.biomedcentral.com; 2012;13: R77.
Abstract
We introduce ggbio, a new methodology to visualize and explore genomics annotationsand high-throughput data. The plots provide detailed views of genomic regions,summary views of sequence alignments and splicing patterns, and genome-wide overviewswith karyogram, circular and grand linear layouts. The methods leverage thestatistical functionality available in R, the grammar of graphics and the datahandling capabilities of the Bioconductor project. The plots are specified within amodular framework that enables users to construct plots in a systematic way, and aregenerated directly from Bioconductor data structures. The ggbio R package isavailable athttp://www.bioconductor.org/packages/2.11/bioc/html/ggbio.html.