Gepard
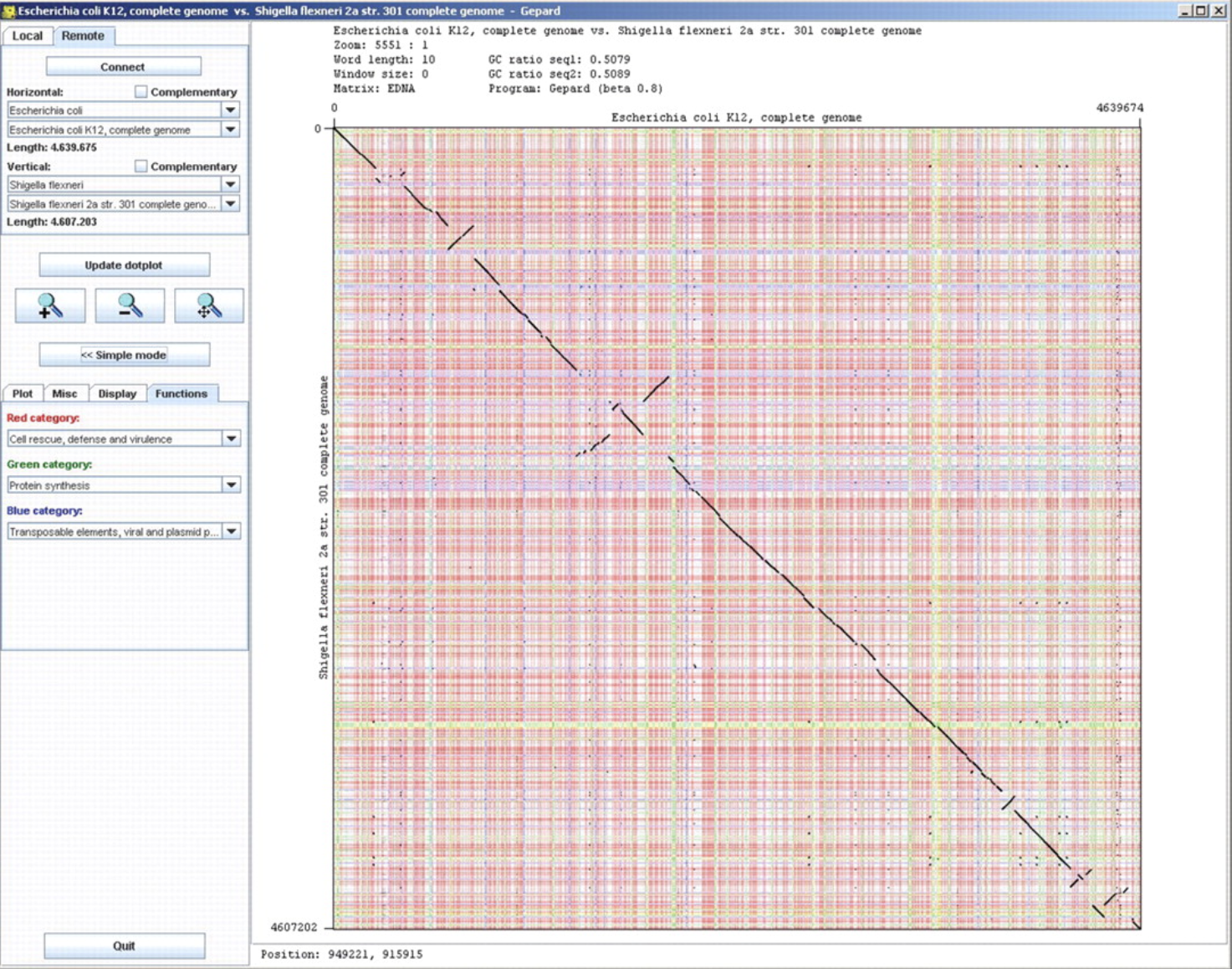 https://academic.oup.com/view-large/figure/1970702/btm039f1.jpeg
https://academic.oup.com/view-large/figure/1970702/btm039f1.jpeg
Methods
linear single view single scale single focus contiguous no abstraction linear orthogonal arrangement between interconnection segment contiguous type segment sparse typeTool
| Access Format | standalone app |
| Supported Files | fasta |
| License | MIT |
| Tool name | Gepard |
| Tool Link | http://mips.gsf.de/services/analysis/gepard |
| Documentation | https://github.com/univieCUBE/gepard/blob/master/tutorial.html |
Paper
Gepard: a rapid and sensitive tool for creating dotplots on genome scale.
Krumsiek J, Arnold R, Rattei T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. academic.oup.com; 2007;23: 1026–1028.
Abstract
Gepard provides a user-friendly, interactive application for the quick creation of dotplots. It utilizes suffix arrays to reduce the time complexity of dotplot calculation to Theta(m*log n). A client-server mode, which is a novel feature for dotplot creation software, allows the user to calculate dotplots and color them by functional annotation without any prior downloading of sequence or annotation data. AVAILABILITY: Both source codes and executable binaries are available at http://mips.gsf.de/services/analysis/gepard