GBrowse_syn
 http://gmod.org/wiki/GBrowse_syn
http://gmod.org/wiki/GBrowse_syn
Methods
linear multiple view multiple scale multiple focus segregated no abstraction linear parallel arrangement between interconnection segment sparse typeTool
| Access Format | web application |
| Supported Files | fasta other |
| License | GPL, Artistic License |
| Tool name | GBrowse_syn |
| Tool Link | http://gmod.org/wiki/GBrowse_syn |
| Documentation | http://gmod.org/wiki/GBrowse_syn |
Paper
Using the Generic Synteny Browser (GBrowse_syn)
McKay SJ, Vergara IA, Stajich JE. Using the Generic Synteny Browser (GBrowse_syn). Curr Protoc Bioinformatics. Wiley Online Library; 2010;Chapter 9: Unit 9.12.
Abstract
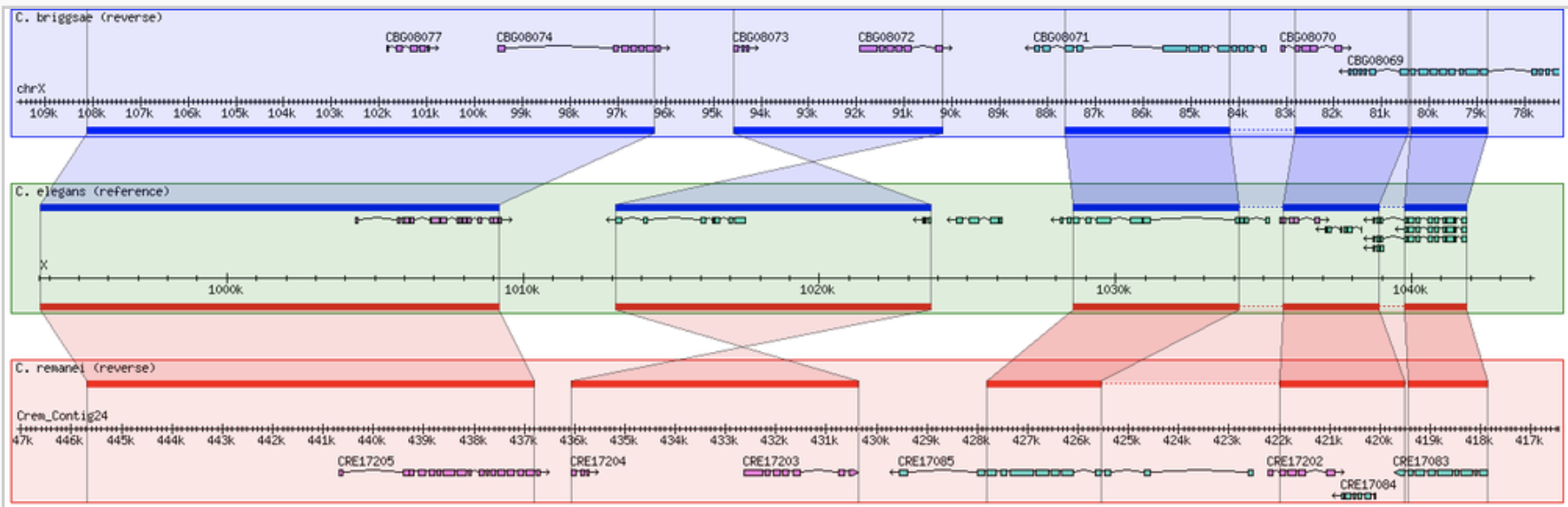
Genome Browsers are software that allow the user to view genome annotations in the context of a reference sequence, such as a chromosome, contig, scaffold, etc. The Generic Genome Browser (GBrowse) is an open source genome browser package developed as part of the Generic Model Database Project (see Unit 9.9; Stein et at., 2002). The increasing number of sequenced genomes has to a corresponding growth in the field of comparative genomics, which requires methods to view and compare multiple genomes. Using the same software framework as GBrowse, the Generic Synteny Browser (GBrowse_syn) allows the comparison of co-linear regions of multiple genomes using the familiar GBrowse-style web page. Like GBrowse, GBrowse_syn can be configured to display any organism and is currently the synteny browser used for model organisms such as C. elegans (WormBase; www.wormbase.org; see Unit 1.8) and Arabidopsis (TAIR; www.arabidopsis.org; see Unit 1.11). GBrowse_syn is part of the GBrowse software package and can be downloaded from the web and run on any unix-like operating system, such as Linux, Solaris, Mac OS X etc. GBrowse_syn is still under active development. This unit will cover installation and configuration as part of the current stable version of GBrowse (v1.71).