Galaxy HiCExplorer
 https://hicexplorer.usegalaxy.eu/
https://hicexplorer.usegalaxy.eu/
Methods
linear single view single scale single focus contiguous no abstraction linear orthogonal arrangement within interconnection segment sparse type segment contiguous type point sparse type point contiguous typeTool
| Access Format | web application |
| Supported Files | fasta |
| License | MIT |
| Tool name | Galaxy HiCExplorer |
| Tool Link | https://hicexplorer.usegalaxy.eu/ |
| Documentation | https://hicexplorer.usegalaxy.eu/tours/core.galaxy_ui |
Paper
Galaxy HiCExplorer: a web server for reproducible Hi-C data analysis, quality control and visualization.
Wolff J, Bhardwaj V, Nothjunge S, Richard G, Renschler G, Gilsbach R, et al. Galaxy HiCExplorer: a web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res. academic.oup.com; 2018;46: W11–W16.
Abstract
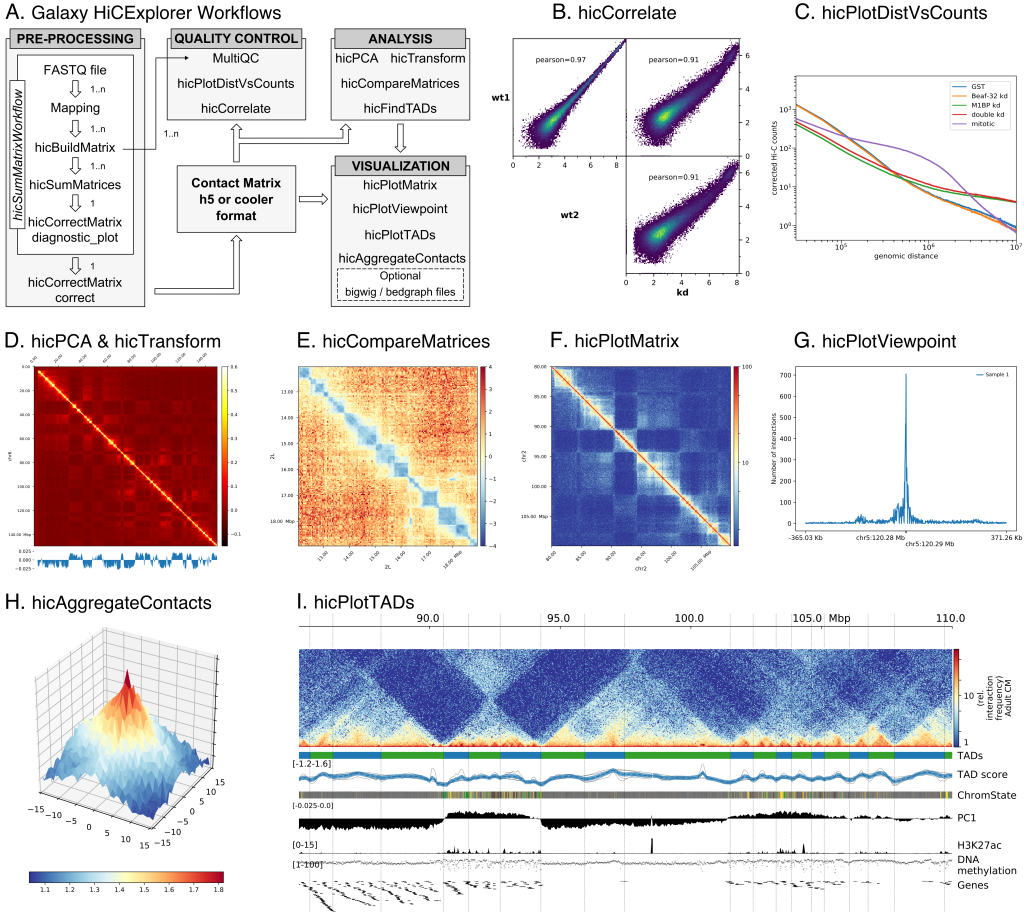
Galaxy HiCExplorer is a web server that facilitates the study of the 3D conformation of chromatin by allowing Hi-C data processing, analysis and visualization. With the Galaxy HiCExplorer web server, users with little bioinformatic background can perform every step of the analysis in one workflow: mapping of the raw sequence data, creation of Hi-C contact matrices, quality assessment, correction of contact matrices and identification of topological associated domains (TADs) and A/B compartments. Users can create publication ready plots of the contact matrix, A/B compartments, and TADs on a selected genomic locus, along with additional information like gene tracks or ChIP-seq signals. Galaxy HiCExplorer is freely usable at: https://hicexplorer.usegalaxy.eu and is available as a Docker container: https://github.com/deeptools/docker-galaxy-hicexplorer.