EnrichedHeatmap
 https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-018-4625-x/figures/2
https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-018-4625-x/figures/2
Methods
linear multiple view single scale single focus segregated no abstraction no arrangement no interconnection point contiguous typeTool
| Access Format | programming language |
| Supported Files | other |
| License | MIT |
| Tool name | EnrichedHeatmap |
| Tool Link | https://bioconductor.org/packages/release/bioc/html/EnrichedHeatmap.html |
| Documentation | https://bioconductor.org/packages/release/bioc/vignettes/EnrichedHeatmap/inst/doc/EnrichedHeatmap.html |
Paper
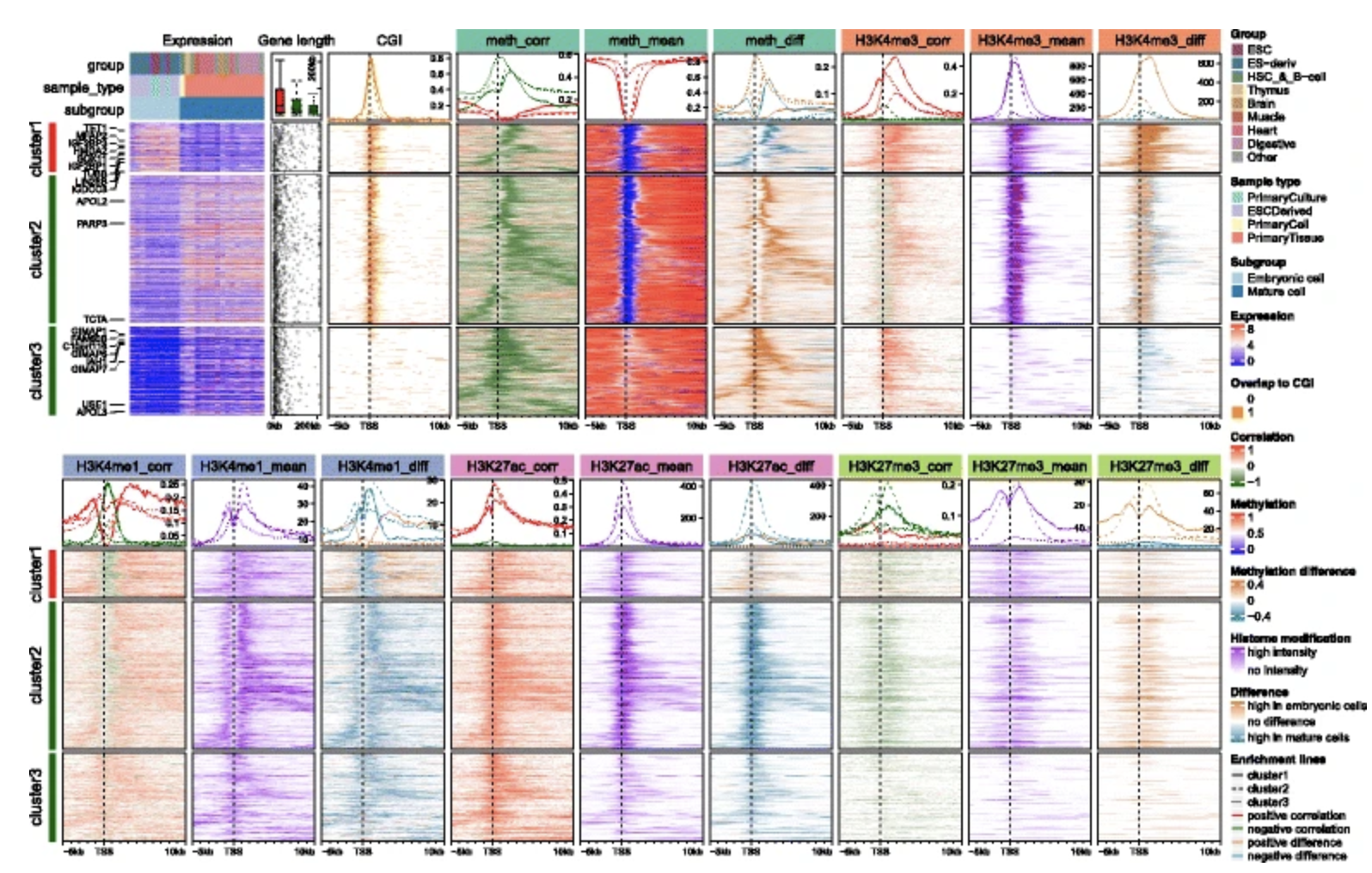
EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations
Gu Z, Eils R, Schlesner M, Ishaque N. EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Genomics. bmcgenomics.biomedcentral.com; 2018;19: 234.
Abstract
Background: High-throughput sequencing data are dramatically increasing in volume. Thus, there is urgent need for efficient tools to perform fast and integrative analysis of multiple data types. Enriched heatmap is a specific form of heatmap that visualizes how genomic signals are enriched over specific target regions. It is commonly used and efficient at revealing enrichment patterns especially for high dimensional genomic and epigenomic datasets. Results: We present a new R package named EnrichedHeatmap that efficiently visualizes genomic signal enrichment. It provides advanced solutions for normalizing genomic signals within target regions as well as offering highly customizable visualizations. The major advantage of EnrichedHeatmap is the ability to conveniently generate parallel heatmaps as well as complex annotations, which makes it easy to integrate and visualize comprehensive overviews of the patterns and associations within and between complex datasets. Conclusions: EnrichedHeatmap facilitates comprehensive understanding of high dimensional genomic and epigenomic data. The power of EnrichedHeatmap is demonstrated by visualization of the complex associations between DNA methylation, gene expression and various histone modifications.